dbAMP 2.0: updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data
- PMID: 34850155
- PMCID: PMC8690246
- DOI: 10.1093/nar/gkab1080
dbAMP 2.0: updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data
Abstract
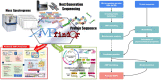
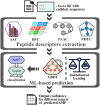
The last 18 months, or more, have seen a profound shift in our global experience, with many of us navigating a once-in-100-year pandemic. To date, COVID-19 remains a life-threatening pandemic with little to no targeted therapeutic recourse. The discovery of novel antiviral agents, such as vaccines and drugs, can provide therapeutic solutions to save human beings from severe infections; however, there is no specifically effective antiviral treatment confirmed for now. Thus, great attention has been paid to the use of natural or artificial antimicrobial peptides (AMPs) as these compounds are widely regarded as promising solutions for the treatment of harmful microorganisms. Given the biological significance of AMPs, it was obvious that there was a significant need for a single platform for identifying and engaging with AMP data. This led to the creation of the dbAMP platform that provides comprehensive information about AMPs and facilitates their investigation and analysis. To date, the dbAMP has accumulated 26 447 AMPs and 2262 antimicrobial proteins from 3044 organisms using both database integration and manual curation of >4579 articles. In addition, dbAMP facilitates the evaluation of AMP structures using I-TASSER for automated protein structure prediction and structure-based functional annotation, providing predictive structure information for clinical drug development. Next-generation sequencing (NGS) and third-generation sequencing have been applied to generate large-scale sequencing reads from various environments, enabling greatly improved analysis of genome structure. In this update, we launch an efficient online tool that can effectively identify AMPs from genome/metagenome and proteome data of all species in a short period. In conclusion, these improvements promote the dbAMP as one of the most abundant and comprehensively annotated resources for AMPs. The updated dbAMP is now freely accessible at http://awi.cuhk.edu.cn/dbAMP.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
dbAMP 3.0: updated resource of antimicrobial activity and structural annotation of peptides in the post-pandemic era.Nucleic Acids Res. 2025 Jan 6;53(D1):D364-D376. doi: 10.1093/nar/gkae1019. Nucleic Acids Res. 2025. PMID: 39540425 Free PMC article.
-
dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data.Nucleic Acids Res. 2019 Jan 8;47(D1):D285-D297. doi: 10.1093/nar/gky1030. Nucleic Acids Res. 2019. PMID: 30380085 Free PMC article.
-
DRAMP 3.0: an enhanced comprehensive data repository of antimicrobial peptides.Nucleic Acids Res. 2022 Jan 7;50(D1):D488-D496. doi: 10.1093/nar/gkab651. Nucleic Acids Res. 2022. PMID: 34390348 Free PMC article.
-
Antimicrobial Peptides: An Update on Classifications and Databases.Int J Mol Sci. 2021 Oct 28;22(21):11691. doi: 10.3390/ijms222111691. Int J Mol Sci. 2021. PMID: 34769122 Free PMC article. Review.
-
A review on the diversity of antimicrobial peptides and genome mining strategies for their prediction.Biochimie. 2024 Dec;227(Pt A):99-115. doi: 10.1016/j.biochi.2024.06.013. Epub 2024 Jun 27. Biochimie. 2024. PMID: 38944107 Review.
Cited by
-
ACVPICPred: Inhibitory activity prediction of anti-coronavirus peptides based on artificial neural network.Comput Struct Biotechnol J. 2024 Oct 2;23:3625-3633. doi: 10.1016/j.csbj.2024.09.015. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39469670 Free PMC article.
-
Heterologous Production of Antimicrobial Peptides: Notes to Consider.Protein J. 2024 Apr;43(2):129-158. doi: 10.1007/s10930-023-10174-w. Epub 2024 Jan 5. Protein J. 2024. PMID: 38180586
-
Antimicrobial Peptides: From Design to Clinical Application.Antibiotics (Basel). 2022 Mar 6;11(3):349. doi: 10.3390/antibiotics11030349. Antibiotics (Basel). 2022. PMID: 35326812 Free PMC article. Review.
-
Machine Learning in Antibacterial Drug Design.Front Pharmacol. 2022 May 3;13:864412. doi: 10.3389/fphar.2022.864412. eCollection 2022. Front Pharmacol. 2022. PMID: 35592425 Free PMC article. Review.
-
Ensemble Machine Learning and Predicted Properties Promote Antimicrobial Peptide Identification.Interdiscip Sci. 2024 Dec;16(4):951-965. doi: 10.1007/s12539-024-00640-z. Epub 2024 Jul 7. Interdiscip Sci. 2024. PMID: 38972032
References
-
- Li W., Separovic F., O’Brien-Simpson N.M., Wade J.D.. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021; 50:4932–4973. - PubMed
-
- Chung C.R., Kuo T.R., Wu L.C., Lee T.Y., Horng J.T.. Characterization and identification of antimicrobial peptides with different functional activities. Brief. Bioinform. 2019; 21:1098–1114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 32070659/National Natural Science Foundation of China
- 2021A1515012447/Guangdong Province Basic and Applied Basic Research Fund
- 2021E007/Ganghong Young Scholar Development Fund
- JCYJ20200109150003938/Science, Technology and Innovation Commission of Shenzhen Municipality
- P2-2021-ZYL-001-A/Chinese University of Hong Kong
LinkOut - more resources
Full Text Sources

