Cellular heterogeneity in DNA alkylation repair increases population genetic plasticity
- PMID: 34850170
- PMCID: PMC8643705
- DOI: 10.1093/nar/gkab1143
Cellular heterogeneity in DNA alkylation repair increases population genetic plasticity
Abstract
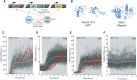
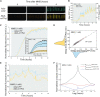
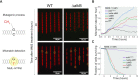
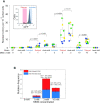
DNA repair mechanisms fulfil a dual role, as they are essential for cell survival and genome maintenance. Here, we studied how cells regulate the interplay between DNA repair and mutation. We focused on the adaptive response that increases the resistance of Escherichia coli cells to DNA alkylation damage. Combination of single-molecule imaging and microfluidic-based single-cell microscopy showed that noise in the gene activation timing of the master regulator Ada is accurately propagated to generate a distinct subpopulation of cells in which all proteins of the adaptive response are essentially absent. Whereas genetic deletion of these proteins causes extreme sensitivity to alkylation stress, a temporary lack of expression is tolerated and increases genetic plasticity of the whole population. We demonstrated this by monitoring the dynamics of nascent DNA mismatches during alkylation stress as well as the frequency of fixed mutations that are generated by the distinct subpopulations of the adaptive response. We propose that stochastic modulation of DNA repair capacity by the adaptive response creates a viable hypermutable subpopulation of cells that acts as a source of genetic diversity in a clonal population.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Stochastic activation of a DNA damage response causes cell-to-cell mutation rate variation.Science. 2016 Mar 4;351(6277):1094-7. doi: 10.1126/science.aac9786. Science. 2016. PMID: 26941321 Free PMC article.
-
Ada protein- and sequence context-dependent mutagenesis of alkyl phosphotriester lesions in Escherichia coli cells.J Biol Chem. 2020 Jun 26;295(26):8775-8783. doi: 10.1074/jbc.RA120.013657. Epub 2020 May 7. J Biol Chem. 2020. PMID: 32381504 Free PMC article.
-
Ada response - a strategy for repair of alkylated DNA in bacteria.FEMS Microbiol Lett. 2014 Jun;355(1):1-11. doi: 10.1111/1574-6968.12462. Epub 2014 Jun 6. FEMS Microbiol Lett. 2014. PMID: 24810496 Free PMC article. Review.
-
[Chemically methylated DNA repair in Escherichia coli--the role of alkB protein].Postepy Biochem. 2006;52(3):239-46. Postepy Biochem. 2006. PMID: 17201058 Review. Polish.
-
Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage.Nature. 2002 Sep 12;419(6903):174-8. doi: 10.1038/nature00908. Nature. 2002. PMID: 12226667
Cited by
-
Adaptation delay causes a burst of mutations in bacteria responding to oxidative stress.EMBO Rep. 2023 Jan 9;24(1):e55640. doi: 10.15252/embr.202255640. Epub 2022 Nov 17. EMBO Rep. 2023. PMID: 36397732 Free PMC article.
-
Estimating mutation rates under heterogeneous stress responses.PLoS Comput Biol. 2024 May 28;20(5):e1012146. doi: 10.1371/journal.pcbi.1012146. eCollection 2024 May. PLoS Comput Biol. 2024. PMID: 38805543 Free PMC article.
-
Phenotypic heterogeneity in the bacterial oxidative stress response is driven by cell-cell interactions.Cell Rep. 2023 Mar 28;42(3):112168. doi: 10.1016/j.celrep.2023.112168. Epub 2023 Feb 26. Cell Rep. 2023. PMID: 36848288 Free PMC article.
-
Microfluidics for long-term single-cell time-lapse microscopy: Advances and applications.Front Bioeng Biotechnol. 2022 Oct 12;10:968342. doi: 10.3389/fbioe.2022.968342. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36312536 Free PMC article. Review.
-
Phenotype switching of the mutation rate facilitates adaptive evolution.Genetics. 2023 Aug 31;225(1):iyad111. doi: 10.1093/genetics/iyad111. Genetics. 2023. PMID: 37293818 Free PMC article.
References
-
- Friedberg E.C., Walker G.C., Siede W., Wood R.D.. DNA Repair and Mutagenesis. 2005; USA: American Society for Microbiology Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

