Establishment of patient-derived organoid models of lower-grade glioma
- PMID: 34850183
- PMCID: PMC8972292
- DOI: 10.1093/neuonc/noab273
Establishment of patient-derived organoid models of lower-grade glioma
Abstract
Background: Historically, creating patient-derived models of lower-grade glioma (LGG) has been challenging, contributing to few experimental platforms that support laboratory-based investigations of this disease. Although organoid modeling approaches have recently been employed to create in vitro models of high-grade glioma (HGG), it is unknown whether this approach can be successfully applied to LGG.
Methods: In this study, we developed an optimized protocol for the establishment of organoids from LGG primary tissue samples by utilizing physiologic (5%) oxygenation conditions and employed it to produce the first known suite of these models. To assess their fidelity, we surveyed key biological features of patient-derived organoids using metabolic, genomic, histologic, and lineage marker gene expression assays.
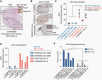
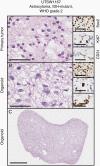
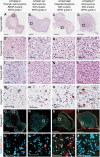
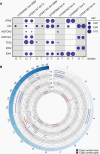
Results: Organoid models were created with a success rate of 91% (n = 20/22) from primary tumor samples across glioma histological subtypes and tumor grades (WHO Grades 1-4), and a success rate of 87% (13/15) for WHO Grade 1-3 tumors. Patient-derived organoids recapitulated stemness, proliferative, and tumor-stromal composition profiles of their respective parental tumor specimens. Cytoarchitectural, mutational, and metabolic traits of parental tumors were also conserved. Importantly, LGG organoids were maintained in vitro for weeks to months and reanimated after biobanking without loss of integrity.
Conclusions: We report an efficient method for producing faithful in vitro models of LGG. New experimental platforms generated through this approach are well positioned to support preclinical studies of this disease, particularly those related to tumor immunology, tumor-stroma interactions, identification of novel drug targets, and personalized assessments of treatment response profiles.
Keywords: glioblastoma; glioma; lower grade glioma; organoids; preclinical models.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Figures

References
-
- Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. - PubMed
-
- Hottinger AF, Abdullah KG, Stupp R. Current standards of care in glioblastoma therapy. In: Glioblastoma. Elsevier Inc.; 2016: 73– 80. doi: 10.1016/B978-0-323-47660-7.00006-9 - DOI
-
- Post GR, Dawson G. Characterization of a cell line derived from a human oligodendroglioma. Mol Chem Neuropathol. 1992;16(3):303–317. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

