FGF7-FGFR2 autocrine signaling increases growth and chemoresistance of fusion-positive rhabdomyosarcomas
- PMID: 34850536
- PMCID: PMC8936514
- DOI: 10.1002/1878-0261.13145
FGF7-FGFR2 autocrine signaling increases growth and chemoresistance of fusion-positive rhabdomyosarcomas
Abstract
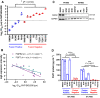
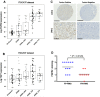
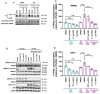
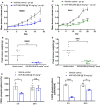
Rhabdomyosarcomas are aggressive pediatric soft-tissue sarcomas and include high-risk PAX3-FOXO1 fusion-gene-positive cases. Fibroblast growth factor receptor 4 (FGFR4) is known to contribute to rhabdomyosarcoma progression; here, we sought to investigate the involvement and potential for therapeutic targeting of other FGFRs in this disease. Cell-based screening of FGFR inhibitors with potential for clinical repurposing (NVP-BGJ398, nintedanib, dovitinib, and ponatinib) revealed greater sensitivity of fusion-gene-positive versus fusion-gene-negative rhabdomyosarcoma cell lines and was shown to be correlated with high expression of FGFR2 and its specific ligand, FGF7. Furthermore, patient samples exhibit higher mRNA levels of FGFR2 and FGF7 in fusion-gene-positive versus fusion-gene-negative rhabdomyosarcomas. Sustained intracellular mitogen-activated protein kinase (MAPK) activity and FGF7 secretion into culture media during serum starvation of PAX3-FOXO1 rhabdomyosarcoma cells together with decreased cell viability after genetic silencing of FGFR2 or FGF7 was in keeping with a novel FGF7-FGFR2 autocrine loop. FGFR inhibition with NVP-BGJ398 reduced viability and was synergistic with SN38, the active metabolite of irinotecan. In vivo, NVP-BGJ398 abrogated xenograft growth and warrants further investigation in combination with irinotecan as a therapeutic strategy for fusion-gene-positive rhabdomyosarcomas.
Keywords: FGF7; FGFR2; NVP-BGJ398; autocrine loop; fibroblast growth factor receptor; rhabdomyosarcoma.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;2:116–29. - PubMed
-
- Dell'Era P, Ronca R, Coco L, Nicoli S, Metra M, Presta M. Fibroblast growth factor receptor‐1 is essential for in vitro cardiomyocyte development. Circ Res. 2003;5:414–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

