Addressing noise in co-expression network construction
- PMID: 34850822
- PMCID: PMC8769892
- DOI: 10.1093/bib/bbab495
Addressing noise in co-expression network construction
Abstract
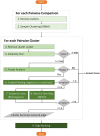
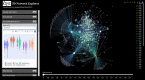
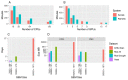
Gene co-expression networks (GCNs) provide multiple benefits to molecular research including hypothesis generation and biomarker discovery. Transcriptome profiles serve as input for GCN construction and are derived from increasingly larger studies with samples across multiple experimental conditions, treatments, time points, genotypes, etc. Such experiments with larger numbers of variables confound discovery of true network edges, exclude edges and inhibit discovery of context (or condition) specific network edges. To demonstrate this problem, a 475-sample dataset is used to show that up to 97% of GCN edges can be misleading because correlations are false or incorrect. False and incorrect correlations can occur when tests are applied without ensuring assumptions are met, and pairwise gene expression may not meet test assumptions if the expression of at least one gene in the pairwise comparison is a function of multiple confounding variables. The 'one-size-fits-all' approach to GCN construction is therefore problematic for large, multivariable datasets. Recently, the Knowledge Independent Network Construction toolkit has been used in multiple studies to provide a dynamic approach to GCN construction that ensures statistical tests meet assumptions and confounding variables are addressed. Additionally, it can associate experimental context for each edge of the network resulting in context-specific GCNs (csGCNs). To help researchers recognize such challenges in GCN construction, and the creation of csGCNs, we provide a review of the workflow.
Keywords: co-expression; gene expression; multidimensional; networks; noise.
© The Author(s) 2021. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

