Genome Sequencing Identifies Previously Unrecognized Klebsiella pneumoniae Outbreaks in Neonatal Intensive Care Units in the Philippines
- PMID: 34850834
- PMCID: PMC8634409
- DOI: 10.1093/cid/ciab776
Genome Sequencing Identifies Previously Unrecognized Klebsiella pneumoniae Outbreaks in Neonatal Intensive Care Units in the Philippines
Abstract
Background: Klebsiella pneumoniae is a critically important pathogen in the Philippines. Isolates are commonly resistant to at least 2 classes of antibiotics, yet mechanisms and spread of its resistance are not well studied.
Methods: A retrospective sequencing survey was performed on carbapenem-, extended spectrum beta-lactam-, and cephalosporin-resistant Klebsiella pneumoniae isolated at 20 antimicrobial resistance (AMR) surveillance sentinel sites from 2015 through 2017. We characterized 259 isolates using biochemical methods, antimicrobial susceptibility testing, and whole-genome sequencing (WGS). Known AMR mechanisms were identified. Potential outbreaks were investigated by detecting clusters from epidemiologic, phenotypic, and genome-derived data.
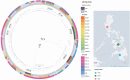
Results: Prevalent AMR mechanisms detected include blaCTX-M-15 (76.8%) and blaNDM-1 (37.5%). An epidemic IncFII(Yp) plasmid carrying blaNDM-1 was also detected in 46 isolates from 6 sentinel sites and 14 different sequence types (STs). This plasmid was also identified as the main vehicle of carbapenem resistance in 2 previously unrecognized local outbreaks of ST348 and ST283 at 2 different sentinel sites. A third local outbreak of ST397 was also identified but without the IncFII(Yp) plasmid. Isolates in each outbreak site showed identical STs and K- and O-loci, and similar resistance profiles and AMR genes. All outbreak isolates were collected from blood of children aged < 1 year.
Conclusion: WGS provided a better understanding of the epidemiology of multidrug resistant Klebsiella in the Philippines, which was not possible with only phenotypic and epidemiologic data. The identification of 3 previously unrecognized Klebsiella outbreaks highlights the utility of WGS in outbreak detection, as well as its importance in public health and in implementing infection control programs.
Keywords: K. pneumoniae; antimicrobial resistance; outbreak detection; whole genome sequencing.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures

References
-
- O’ Neil J. Review on Antibiotic resistance. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance, 2014. Available at: https://amr-review.org/sites/default/files/AMR Review Paper - Tackling a.... Accessed 10 June 2021.
-
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS): molecular methods for antimicrobial resistance (AMR) diagnostics to enhance the Global Antimicrobial Resistance Surveillance System. No. WHO/WSI/AMR/2019.1. Geneva: World Health Organization, 2019.
-
- World Health Organization. WHONET software. Geneva: WHO, 2021.

