Clones and Clusters of Antimicrobial-Resistant Klebsiella From Southwestern Nigeria
- PMID: 34850837
- PMCID: PMC8634535
- DOI: 10.1093/cid/ciab769
Clones and Clusters of Antimicrobial-Resistant Klebsiella From Southwestern Nigeria
Abstract
Background: Klebsiella pneumoniae is a World Health Organization high-priority antibiotic-resistant pathogen. However, little is known about Klebsiella lineages circulating in Nigeria.
Methods: We performed whole-genome sequencing (WGS) of 141 Klebsiella isolated between 2016 and 2018 from clinical specimens at 3 antimicrobial-resistance (AMR) sentinel surveillance tertiary hospitals in southwestern Nigeria. We conducted in silico multilocus sequence typing; AMR gene, virulence gene, plasmid, and K and O loci profiling; as well as phylogenetic analyses, using publicly available tools and Nextflow pipelines.
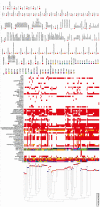
Results: Phylogenetic analysis revealed that the majority of the 134 K. pneumoniae and 5 K. quasipneumoniae isolates from Nigeria characterized are closely related to globally disseminated multidrug-resistant clones. Of the 39 K. pneumoniae sequence types (STs) identified, the most common were ST307 (15%), ST5241 (12%), ST15 (~9%), and ST25 (~6%). ST5241, 1 of 10 novel STs detected, is a single locus variant of ST636 carrying dfrA14, tetD, qnrS, and oqxAB resistance genes. The extended-spectrum β-lactamase (ESBL) gene blaCTX_M-15 was seen in 72% of K. pneumoniae genomes, while 8% encoded a carbapenemase. No isolate carried a combination of carbapenemase-producing genes. Four likely outbreak clusters from 1 facility, within STs 17, 25, 307, and 5241, were ESBL but not carbapenemase-bearing clones.
Conclusions: This study uncovered known and novel K. pneumoniae lineages circulating in 3 hospitals in Southwest Nigeria that include multidrug-resistant ESBL producers. Carbapenemase-producing isolates remain uncommon. WGS retrospectively identified outbreak clusters, pointing to the value of genomic approaches in AMR surveillance for improving infection prevention and control in Nigerian hospitals.
Keywords: Klebsiella; Nigeria; antimicrobial resistance; genomic surveillance.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures

References
-
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 2016; 24:944-56. - PubMed
-
- Olalekan A, Onwugamba F, Iwalokun B, Mellmann A, Becker K, Schaumburg F. High proportion of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae among extended-spectrum β-lactamase-producers in Nigerian hospitals. J Glob Antimicrob Resist 2020; 21:8-12. - PubMed
-
- Müller-Schulte E, Tuo MN, Akoua-Koffi C, Schaumburg F, Becker SL. High prevalence of ESBL-producing Klebsiella pneumoniae in clinical samples from central Côte d’Ivoire. Int J Infect Dis 2020; 91:207-9. - PubMed

