Overcoming Data Bottlenecks in Genomic Pathogen Surveillance
- PMID: 34850839
- PMCID: PMC8634317
- DOI: 10.1093/cid/ciab785
Overcoming Data Bottlenecks in Genomic Pathogen Surveillance
Abstract
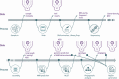
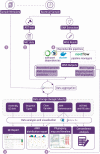
Performing whole genome sequencing (WGS) for the surveillance of antimicrobial resistance offers the ability to determine not only the antimicrobials to which rates of resistance are increasing, but also the evolutionary mechanisms and transmission routes responsible for the increase at local, national, and global scales. To derive WGS-based outputs, a series of processes are required, beginning with sample and metadata collection, followed by nucleic acid extraction, library preparation, sequencing, and analysis. Throughout this pathway there are many data-related operations required (informatics) combined with more biologically focused procedures (bioinformatics). For a laboratory aiming to implement pathogen genomics, the informatics and bioinformatics activities can be a barrier to starting on the journey; for a laboratory that has already started, these activities may become overwhelming. Here we describe these data bottlenecks and how they have been addressed in laboratories in India, Colombia, Nigeria, and the Philippines, as part of the National Institute for Health Research Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance. The approaches taken include the use of reproducible data parsing pipelines and genome sequence analysis workflows, using technologies such as Data-flo, the Nextflow workflow manager, and containerization of software dependencies. By overcoming barriers to WGS implementation in countries where genome sampling for some species may be underrepresented, a body of evidence can be built to determine the concordance of antimicrobial sensitivity testing and genome-derived resistance, and novel high-risk clones and unknown mechanisms of resistance can be discovered.
Keywords: WGS; antimicrobial resistance; bioinformatics; metadata; whole genome sequencing.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures

References
-
- World Health Organization. GLASS whole-genome sequencing for surveillance of antimicrobial resistance. 2020. Available at: https://www.who.int/publications/i/item/9789240011007. Accessed 29 September 2021.
-
- Ellington MJ, Ekelund O, Aarestrup FM, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST subcommittee. Clin Microbiol Infect 2017; 23:2–22. - PubMed
-
- Su M, Satola SW, Read TD. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 2019; 57:e01405-18. Available at: https://jcm.asm.org/content/57/3/e01405-18. Accessed 30 September 2020. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

