Time Series Transcriptomic Analysis of Bronchoalveolar Lavage Cells from Piglets Infected with Virulent or Low-Virulent Porcine Reproductive and Respiratory Syndrome Virus 1
- PMID: 34851149
- PMCID: PMC8826917
- DOI: 10.1128/JVI.01140-21
Time Series Transcriptomic Analysis of Bronchoalveolar Lavage Cells from Piglets Infected with Virulent or Low-Virulent Porcine Reproductive and Respiratory Syndrome Virus 1
Abstract
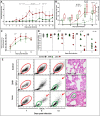
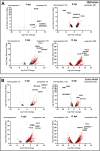
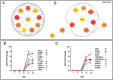
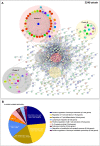
Porcine reproductive and respiratory syndrome virus (PRRSV) has evolved to escape the immune surveillance for a survival advantage leading to a strong modulation of host's immune responses and favoring secondary bacterial infections. However, limited data are available on how the immunological and transcriptional responses elicited by virulent and low-virulent PRRSV-1 strains are comparable and how they are conserved during the infection. To explore the kinetic transcriptional signature associated with the modulation of host immune response at lung level, a time-series transcriptomic analysis was performed in bronchoalveolar lavage cells upon experimental in vivo infection with two PRRSV-1 strains of different virulence, virulent subtype 3 Lena strain or the low-virulent subtype 1 3249 strain. The time-series analysis revealed overlapping patterns of dysregulated genes enriched in T-cell signaling pathways among both virulent and low-virulent strains, highlighting an upregulation of co-stimulatory and co-inhibitory immune checkpoints that were disclosed as Hub genes. On the other hand, virulent Lena infection induced an early and more marked "negative regulation of immune system process" with an overexpression of co-inhibitory receptors genes related to T-cell and NK cell functions, in association with more severe lung lesion, lung viral load, and BAL cell kinetics. These results underline a complex network of molecular mechanisms governing PRRSV-1 immunopathogenesis at lung level, revealing a pivotal role of co-inhibitory and co-stimulatory immune checkpoints in the pulmonary disease, which may have an impact on T-cell activation and related pathways. These immune checkpoints, together with the regulation of cytokine-signaling pathways, modulated in a virulence-dependent fashion, orchestrate an interplay among pro- and anti-inflammatory responses. IMPORTANCE Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the major threats to swine health and global production, causing substantial economic losses. We explore the mechanisms involved in the modulation of host immune response at lung level performing a time-series transcriptomic analysis upon experimental infection with two PRRSV-1 strains of different virulence. A complex network of molecular mechanisms was revealed to control the immunopathogenesis of PRRSV-1 infection, highlighting an interplay among pro- and anti-inflammatory responses as a potential mechanism to restrict inflammation-induced lung injury. Moreover, a pivotal role of co-inhibitory and co-stimulatory immune checkpoints was evidenced, which may lead to progressive dysfunction of T cells, impairing viral clearance and leading to persistent infection, favoring as well secondary bacterial infections or viral rebound. However, further studies should be conducted to evaluate the functional role of immune checkpoints in advanced stages of PRRSV infection and explore a possible T-cell exhaustion state.
Keywords: Hub genes; T-cells; bronchoalveolar lavage cells; immune checkpoints; porcine reproductive and respiratory syndrome virus; time-series transcriptomic analysis; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






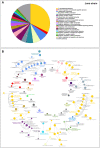
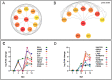
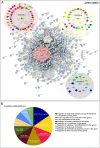
References
-
- Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Wang C, Yeske PE, Mowrer CL, Haley CA. 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Heal Prod 21:72–84.
-
- Meulenberg JJM, Hulst MM, De Meijer EJ, Moonen PLJM, Den Besten A, De Kluyver EP, Wensvoort G, Moormann RJM. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62–72. 10.1006/viro.1993.1008. - DOI - PMC - PubMed
-
- Brinton MA, Gulyaeva A, Balasuriya UBR, Dunowska M, Faaberg KS, Goldberg T, Leung FC, Nauwynck HJ, Snijder EJ, Stadejek T, Gorbalenya AE. 2018. Proposal 2017.012S.A.v1. Expansion of the rank structure of the family Arteriviridae and renaming its taxa. https://talk.ictvonline.org/ICTV/proposals/2017.001S.012-017S.R.Nidovira.... Accessed 15 October 2021.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

