pmrCAB Recombination Events among Colistin-Susceptible and -Resistant Acinetobacter baumannii Clinical Isolates Belonging to International Clone 7
- PMID: 34851165
- PMCID: PMC8636104
- DOI: 10.1128/msphere.00746-21
pmrCAB Recombination Events among Colistin-Susceptible and -Resistant Acinetobacter baumannii Clinical Isolates Belonging to International Clone 7
Abstract
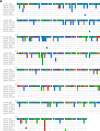
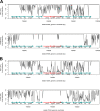
Acinetobacter baumannii is a successful nosocomial pathogen due to its genomic plasticity. Homologous recombination allows genetic exchange and allelic variation among different clonal lineages and is one of the mechanisms associated with horizontal gene transfer (HGT) of resistance determinants. The main mechanism of colistin resistance in A. baumannii is mediated through mutations in the pmrCAB operon. Here, we describe two A. baumannii clinical isolates belonging to International Clone 7 (IC7) that have undergone recombination in the pmrCAB operon and evaluate the contribution of mobile genetic elements (MGE) to this phenomenon. Isolates 67569 and 72554 were colistin susceptible and resistant, respectively, and were submitted for short- and long-read genome sequencing using Illumina MiSeq and MinION platforms. Hybrid assemblies were built with Unicycler, and the assembled genomes were compared to reference genomes using NUCmer, Cortex, and SplitsTree. Genomes were annotated using Prokka, and MGEs were identified with ISfinder and repeat match. Both isolates presented a 21.5-kb recombining region encompassing pmrCAB. In isolate 67659, this region originated from IC5, while in isolate 72554 multiple recombination events might have happened, with the 5-kb recombining region encompassing pmrCAB associated with an isolate representing IC4. We could not identify MGEs involved in the mobilization of pmrCAB in these isolates. In summary, A. baumannii belonging to IC7 can present additional sequence divergence due to homologous recombination across clonal lineages. Such variation does not seem to be driven by antibiotic pressure but could contribute to HGT mediating colistin resistance. IMPORTANCE Colistin resistance rates among Acinetobacter baumannii clinical isolates have increased over the last 20 years. Despite reports of the spread of plasmid-mediated colistin resistance among Enterobacterales, the presence of mcr-type genes in Acinetobacter spp. remains rare, and reduced colistin susceptibility is mainly associated with the acquisition of nonsynonymous mutations in pmrCAB. We have recently demonstrated that distinct pmrCAB sequences are associated with different A. baumannii International Clones (IC). In this study, we identified the presence of homologous recombination as an additional cause of genetic variation in this operon, which, to the best of our knowledge, was not mediated by mobile genetic elements. Even though this phenomenon was observed in both colistin-susceptible and -resistant isolates, it has the potential to contribute to the spread of resistance-conferring alleles, leading to reduced susceptibility to this last-resort antimicrobial agent.
Keywords: Gram-negative bacilli; colistin resistance; insertion sequences; mobile genetic elements; polymyxins.
Conflict of interest statement
The authors declare a conflict of interest.
Figures



Similar articles
-
Diversity of amino acid substitutions in PmrCAB associated with colistin resistance in clinical isolates of Acinetobacter baumannii.Int J Antimicrob Agents. 2020 Mar;55(3):105862. doi: 10.1016/j.ijantimicag.2019.105862. Epub 2019 Dec 16. Int J Antimicrob Agents. 2020. PMID: 31837449
-
Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates.mBio. 2019 Jul 16;10(4):e01083-19. doi: 10.1128/mBio.01083-19. mBio. 2019. PMID: 31311879 Free PMC article.
-
A novel mutation in pmrB mediates colistin resistance during therapy of Acinetobacter baumannii.Int J Antimicrob Agents. 2017 Jun;49(6):727-733. doi: 10.1016/j.ijantimicag.2017.01.031. Epub 2017 Apr 21. Int J Antimicrob Agents. 2017. PMID: 28438568
-
Colistin Resistance Mechanism and Management Strategies of Colistin-Resistant Acinetobacter baumannii Infections.Pathogens. 2024 Nov 28;13(12):1049. doi: 10.3390/pathogens13121049. Pathogens. 2024. PMID: 39770308 Free PMC article. Review.
-
Colistin Resistance in Acinetobacter baumannii: Basic and Clinical Insights.Biol Pharm Bull. 2025;48(3):213-221. doi: 10.1248/bpb.b23-00642. Biol Pharm Bull. 2025. PMID: 40024691 Review.
Cited by
-
Portrait of a killer: Uncovering resistance mechanisms and global spread of Acinetobacter baumannii.PLoS Pathog. 2023 Aug 10;19(8):e1011520. doi: 10.1371/journal.ppat.1011520. eCollection 2023 Aug. PLoS Pathog. 2023. PMID: 37561719 Free PMC article.
-
Genomic epidemiology reveals antibiotic resistance transfer and polyclonal dissemination of Acinetobacter baumannii in a Paraguayan hospital.Antimicrob Agents Chemother. 2025 Aug 6;69(8):e0007725. doi: 10.1128/aac.00077-25. Epub 2025 Jul 8. Antimicrob Agents Chemother. 2025. PMID: 40626882 Free PMC article.
References
-
- Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, Murray PR, Segre JA, NISC Comparative Sequence Program . 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc Natl Acad Sci USA 108:13758–13763. doi:10.1073/pnas.1104404108. - DOI - PMC - PubMed
-
- Müller C, Stefanik D, Wille J, Hackel M, Higgins PG, Seifert H. 2019. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii clinical isolates and identification of the novel international clone IC9: results from a worldwide surveillance study (2012–2016). 29th Eur Cong Clin Microbiol Infect Dis (ECCMID), Amsterdam, Netherlands.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

