Hepatitis B Virus X Protein Expression Is Tightly Regulated by N6-Methyladenosine Modification of Its mRNA
- PMID: 34851655
- PMCID: PMC8865537
- DOI: 10.1128/JVI.01655-21
Hepatitis B Virus X Protein Expression Is Tightly Regulated by N6-Methyladenosine Modification of Its mRNA
Abstract
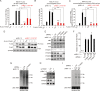
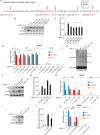
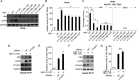
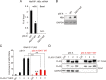
Hepatitis B virus (HBV) encodes a regulatory protein, termed HBx, that has been intensely studied in the past and shown to play a key role(s) in viral transcription and replication. In addition, a huge body of work exists in the literature related to signal transduction and possible mechanism(s) leading to hepatocarcinogenesis associated with infection. We have previously reported that HBV transcripts are modified by N6-methyladenosine (m6A) at the single consensus DRACH motif at nucleotides (nt) 1905 to 1909 in the epsilon structural element, and this m6A modification affects the HBV life cycle. In this study, we present evidence that additional variants of m6A (DRACH) motifs located within nt 1606 to 1809 correspond to the coding region of HBx mRNA and 3' untranslated region (UTR) of other viral mRNAs. Using the mutants of additional m6A sites in nt 1606 to 1809 and a depletion strategy of m6A methyltransferases (METTL3/14) and reader proteins (YTHDFs), we show that m6A modification at nt 1616, located in the HBx coding region, regulates HBx protein expression. The HBx RNA and protein expression levels were notably increased by the silencing of m6A reader YTHDF2 and methyltransferases as well as the mutation of m6A sites in the HBx coding region. However, other viral protein expression levels were not affected by the m6A modification at nt 1616. Thus, m6A modifications in the HBx open reading frame (ORF) downregulate HBx protein expression, commonly seen during HBV transfections, transgenic mice, and natural infections of human hepatocytes. These studies identify the functional role of m6A modification in the subtle regulation of HBx protein expression consistent with its possible role in establishing chronic hepatitis. IMPORTANCE N6-methyladenosien (m6A) modifications recently have been implicated in the HBV life cycle. Previously, we observed that m6A modification occurs in the adenosine at nt 1907 of the HBV genome, and this modification regulates the viral life cycle. Here, we identified an additional m6A site located in nt 1616 of the HBV genome. This modification negatively affects HBx RNA and protein expression. In the absence of m6A methyltransferases (METTL3/14) and reader protein (YTHDF2), the HBx RNA and protein expression were increased. Using HBV mutants that lack m6A in the HBx coding region, we present the unique positional effects of m6A in the regulation of HBx protein expression.
Keywords: HBV life cycle; HBx protein; N6-methyladenosine; hepatitis B virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Yan H, Zhong GC, Xu GW, He WH, Jing ZY, Gao ZC, Huang Y, Qi YH, Peng B, Wang HM, Fu LR, Song M, Chen P, Gao WQ, Ren BJ, Sun YY, Cai T, Feng XF, Sui JH, Li WH. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. 10.7554/eLife.00049. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

