Human Rhinovirus Infection of the Respiratory Tract Affects Sphingolipid Synthesis
- PMID: 34851798
- PMCID: PMC8937237
- DOI: 10.1165/rcmb.2021-0443OC
Human Rhinovirus Infection of the Respiratory Tract Affects Sphingolipid Synthesis
Abstract
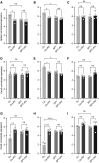
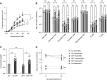
The 17q21 asthma susceptibility locus includes asthma risk alleles associated with decreased sphingolipid synthesis, likely resulting from increased expression of ORMDL3. ORMDL3 inhibits serine-palmitoyl transferase (SPT), the rate-limiting enzyme of de novo sphingolipid synthesis. There is evidence that decreased sphingolipid synthesis is critical to asthma pathogenesis. Children with asthma and 17q21 asthma risk alleles display decreased sphingolipid synthesis in blood cells. Reduced SPT activity results in airway hyperreactivity, a hallmark feature of asthma. 17q21 asthma risk alleles are also linked to childhood infections with human rhinovirus (RV). This study evaluates the interaction of RV with the de novo sphingolipid synthesis pathway, and the alterative effects of concurrent SPT inhibition in SPT-deficient mice and human airway epithelial cells. In mice, RV infection shifted lung sphingolipid synthesis gene expression to a pattern that resembles genetic SPT deficiency, including decreased expression of Sptssa, a small SPT subunit. This pattern was pronounced in lung epithelial cellular adhesion molecule (EpCAM+) cells and reproduced in human bronchial epithelial cells. RV did not affect Sptssa expression in lung CD45+ immune cells. RV increased sphingolipids unique to the de novo synthesis pathway in mouse lung and human airway epithelial cells. Interestingly, these de novo sphingolipid species were reduced in the blood of RV-infected wild-type mice. RV exacerbated SPT deficiency-associated airway hyperreactivity. Airway inflammation was similar in RV-infected wild-type and SPT-deficient mice. This study reveals the effects of RV infection on the de novo sphingolipid synthesis pathway, elucidating a potential mechanistic link between 17q21 asthma risk alleles and rhinoviral infection.
Keywords: 17q21; asthma; early childhood wheeze; rhinovirus; sphingolipids.
Figures




Similar articles
-
Orosomucoid-like 3 Supports Rhinovirus Replication in Human Epithelial Cells.Am J Respir Cell Mol Biol. 2020 Jun;62(6):783-792. doi: 10.1165/rcmb.2019-0237OC. Am J Respir Cell Mol Biol. 2020. PMID: 32078788 Free PMC article.
-
Sphingolipids and Asthma.Adv Exp Med Biol. 2022;1372:145-155. doi: 10.1007/978-981-19-0394-6_10. Adv Exp Med Biol. 2022. PMID: 35503179
-
Airway reactivity and sphingolipids-implications for childhood asthma.Mol Cell Pediatr. 2015 Dec;2(1):13. doi: 10.1186/s40348-015-0025-3. Epub 2015 Dec 4. Mol Cell Pediatr. 2015. PMID: 26637347 Free PMC article.
-
Perinatal origins of chronic lung disease: mechanisms-prevention-therapy-sphingolipid metabolism and the genetic and perinatal origins of childhood asthma.Mol Cell Pediatr. 2021 Dec 20;8(1):22. doi: 10.1186/s40348-021-00130-y. Mol Cell Pediatr. 2021. PMID: 34931265 Free PMC article. Review.
-
Sphingolipids, ORMDL3 and asthma: what is the evidence?Curr Opin Clin Nutr Metab Care. 2017 Mar;20(2):99-103. doi: 10.1097/MCO.0000000000000349. Curr Opin Clin Nutr Metab Care. 2017. PMID: 28030368 Review.
Cited by
-
Neutrophilia in severe asthma is reduced in Ormdl3 overexpressing mice.FASEB J. 2023 Mar;37(3):e22799. doi: 10.1096/fj.202201821R. FASEB J. 2023. PMID: 36753412 Free PMC article.
-
Possible Involvement of Lysophospholipids in Severe Asthma as Novel Lipid Mediators.Biomolecules. 2025 Jan 27;15(2):182. doi: 10.3390/biom15020182. Biomolecules. 2025. PMID: 40001485 Free PMC article. Review.
-
Dietary long-chain omega 3 fatty acids modify sphingolipid metabolism to facilitate airway hyperreactivity.Sci Rep. 2022 Nov 17;12(1):19735. doi: 10.1038/s41598-022-21083-w. Sci Rep. 2022. PMID: 36396956 Free PMC article.
-
Plasma Metabonomics of Human Adenovirus-infected Patients with Pneumonia and Upper Respiratory Tract Infection.Curr Med Sci. 2024 Feb;44(1):121-133. doi: 10.1007/s11596-024-2835-9. Epub 2024 Feb 23. Curr Med Sci. 2024. PMID: 38393525
References
-
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature . 2007;448:470–473. - PubMed
-
- Loss GJ, Depner M, Hose AJ, Genuneit J, Karvonen AM, Hyvärinen A, et al. PASTURE (Protection against Allergy Study in Rural Environments) Study Group The early development of wheeze. Environmental determinants and genetic susceptibility at 17q21. Am J Respir Crit Care Med . 2016;193:889–897. - PubMed
-
- Bisgaard H, Bønnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Møller E, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med . 2009;179:179–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

