Dynamic maximum entropy provides accurate approximation of structured population dynamics
- PMID: 34851948
- PMCID: PMC8668141
- DOI: 10.1371/journal.pcbi.1009661
Dynamic maximum entropy provides accurate approximation of structured population dynamics
Abstract
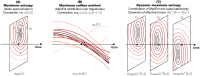
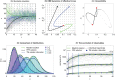
Realistic models of biological processes typically involve interacting components on multiple scales, driven by changing environment and inherent stochasticity. Such models are often analytically and numerically intractable. We revisit a dynamic maximum entropy method that combines a static maximum entropy with a quasi-stationary approximation. This allows us to reduce stochastic non-equilibrium dynamics expressed by the Fokker-Planck equation to a simpler low-dimensional deterministic dynamics, without the need to track microscopic details. Although the method has been previously applied to a few (rather complicated) applications in population genetics, our main goal here is to explain and to better understand how the method works. We demonstrate the usefulness of the method for two widely studied stochastic problems, highlighting its accuracy in capturing important macroscopic quantities even in rapidly changing non-stationary conditions. For the Ornstein-Uhlenbeck process, the method recovers the exact dynamics whilst for a stochastic island model with migration from other habitats, the approximation retains high macroscopic accuracy under a wide range of scenarios in a dynamic environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Jaynes ET. Information theory and statistical mechanics. Phys Rev. 1957. May;106: 620–630. doi: 10.1103/PhysRev.106.620 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

