Towards risk stratification and prediction of disease severity and mortality in COVID-19: Next generation metabolomics for the measurement of host response to COVID-19 infection
- PMID: 34851990
- PMCID: PMC8635335
- DOI: 10.1371/journal.pone.0259909
Towards risk stratification and prediction of disease severity and mortality in COVID-19: Next generation metabolomics for the measurement of host response to COVID-19 infection
Abstract
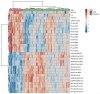
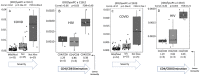
This study investigated the association between COVID-19 infection and host metabolic signatures as prognostic markers for disease severity and mortality. We enrolled 82 patients with RT-PCR confirmed COVID-19 infection who were classified as mild, moderate, or severe/critical based upon their WHO clinical severity score and compared their results with 31 healthy volunteers. Data on demographics, comorbidities and clinical/laboratory characteristics were obtained from medical records. Peripheral blood samples were collected at the time of clinical evaluation or admission and tested by quantitative mass spectrometry to characterize metabolic profiles using selected metabolites. The findings in COVID-19 (+) patients reveal changes in the concentrations of glutamate, valeryl-carnitine, and the ratios of Kynurenine/Tryptophan (Kyn/Trp) to Citrulline/Ornithine (Cit/Orn). The observed changes may serve as predictors of disease severity with a (Kyn/Trp)/(Cit/Orn) Receiver Operator Curve (ROC) AUC = 0.95. Additional metabolite measures further characterized those likely to develop severe complications of their disease, suggesting that underlying immune signatures (Kyn/Trp), glutaminolysis (Glutamate), urea cycle abnormalities (Cit/Orn) and alterations in organic acid metabolism (C5) can be applied to identify individuals at the highest risk of morbidity and mortality from COVID-19 infection. We conclude that host metabolic factors, measured by plasma based biochemical signatures, could prove to be important determinants of Covid-19 severity with implications for prognosis, risk stratification and clinical management.
Conflict of interest statement
R.S. Diaz, M.A. Budib, R. Ayache, R.M.S. Silva, F.C. Silva, S.S. Júnior, H.B.D. Pontes, A.C. Alvarenga, E.C. Arima, W.G. Martins, N.L.F. Silva, M.B. Salzgeber and A.M. Palma have no conflicts of interest to declare. P. D’Amora (consultation: Metabolomycs, Inc.; intellectual property rights (patent pending) Metabolomycs, Inc.; stockholder: Metabolomycs, Inc.). S.S. Evans (stockholder: Metabolomycs, Inc.). I.D.C.G. da Silva (consultation, Metabolomycs, Inc.; intellectual property (patent pending) Metabolomycs, Inc.; board director Metabolomycs, Inc.; stockholder: Metabolomycs, Inc.). R.A. Nagourney (intellectual property rights (patent pending) Metabolomycs, Inc.; board director: Nagourney Institute, board director Metabolomycs, Inc.; stockholder: Metabolomycs, Inc.). Robson Mateus Appel (RMA) was reimbursed for patient registration and sample collection costs by Metabolomycs, Inc. Dr. D’Amora and Dr da Silva of UNIFESP, receive consulting fees from Metabolomycs, Inc. for their cancer-related metabolomic research. We certify that the submission is original work and is not under review at any other publication.
Figures





References
-
- Scarpellini B, Zanoni M, Sucupira MC, Truong HM, Janini LM, da Silva ID, et al.. Correction: Plasma Metabolomics Biosignature According to HIV Stage of Infection, Pace of Disease Progression, Viremia Level and Immunological Response to Treatment. PLoS One. 2017. Feb 24;12(2):e0173164. Erratum for: PLoS One. 2016 Dec 12;11(12):e0161920. doi: 10.1371/journal.pone.0173164 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

