The RNA-binding protein SRSF3 has an essential role in megakaryocyte maturation and platelet production
- PMID: 34852174
- PMCID: PMC8900270
- DOI: 10.1182/blood.2021013826
The RNA-binding protein SRSF3 has an essential role in megakaryocyte maturation and platelet production
Abstract
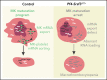
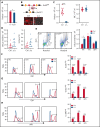
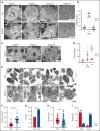
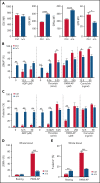
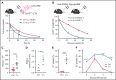
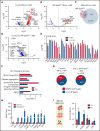
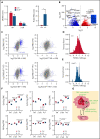
RNA processing is increasingly recognized as a critical control point in the regulation of different hematopoietic lineages including megakaryocytes responsible for the production of platelets. Platelets are anucleate cytoplasts that contain a rich repertoire of RNAs encoding proteins with essential platelet functions derived from the parent megakaryocyte. It is largely unknown how RNA binding proteins contribute to the development and functions of megakaryocytes and platelets. We show that serine-arginine-rich splicing factor 3 (SRSF3) is essential for megakaryocyte maturation and generation of functional platelets. Megakaryocyte-specific deletion of Srsf3 in mice led to macrothrombocytopenia characterized by megakaryocyte maturation arrest, dramatically reduced platelet counts, and abnormally large functionally compromised platelets. SRSF3 deficient megakaryocytes failed to reprogram their transcriptome during maturation and to load platelets with RNAs required for normal platelet function. SRSF3 depletion led to nuclear accumulation of megakaryocyte mRNAs, demonstrating that SRSF3 deploys similar RNA regulatory mechanisms in megakaryocytes as in other cell types. Our study further suggests that SRSF3 plays a role in sorting cytoplasmic megakaryocyte RNAs into platelets and demonstrates how SRSF3-mediated RNA processing forms a central part of megakaryocyte gene regulation. Understanding SRSF3 functions in megakaryocytes and platelets provides key insights into normal thrombopoiesis and platelet pathologies as SRSF3 RNA targets in megakaryocytes are associated with platelet diseases.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures
Similar articles
-
The role of CREG1 in megakaryocyte maturation and thrombocytopoiesis.Int J Biol Sci. 2023 Jul 9;19(11):3614-3627. doi: 10.7150/ijbs.78660. eCollection 2023. Int J Biol Sci. 2023. PMID: 37496998 Free PMC article.
-
Megakaryocyte modification of platelets in thrombocytopenia.Curr Opin Hematol. 2018 Sep;25(5):410-415. doi: 10.1097/MOH.0000000000000451. Curr Opin Hematol. 2018. PMID: 29985173 Review.
-
Inherited thrombocytopenia: novel insights into megakaryocyte maturation, proplatelet formation and platelet lifespan.Platelets. 2016 Sep;27(6):519-25. doi: 10.3109/09537104.2016.1148806. Epub 2016 Mar 30. Platelets. 2016. PMID: 27025194 Free PMC article. Review.
-
Megakaryocyte-specific deletion of the protein-tyrosine phosphatases Shp1 and Shp2 causes abnormal megakaryocyte development, platelet production, and function.Blood. 2013 May 16;121(20):4205-20. doi: 10.1182/blood-2012-08-449272. Epub 2013 Mar 18. Blood. 2013. PMID: 23509158 Free PMC article.
-
Regulation of the genetic code in megakaryocytes and platelets.J Thromb Haemost. 2015 Jun;13 Suppl 1(0 1):S26-32. doi: 10.1111/jth.12965. J Thromb Haemost. 2015. PMID: 26149034 Free PMC article. Review.
Cited by
-
High ploidy large cytoplasmic megakaryocytes are hematopoietic stem cells regulators and essential for platelet production.Nat Commun. 2023 Apr 13;14(1):2099. doi: 10.1038/s41467-023-37780-7. Nat Commun. 2023. PMID: 37055407 Free PMC article.
-
α-Actinin-1 deficiency in megakaryocytes causes low platelet count, platelet dysfunction, and mitochondrial impairment.Blood Adv. 2025 Mar 11;9(5):1185-1201. doi: 10.1182/bloodadvances.2024014805. Blood Adv. 2025. PMID: 39813624 Free PMC article.
-
Oncogenic SRSF3 in health and diseases.Int J Biol Sci. 2023 Jun 12;19(10):3057-3076. doi: 10.7150/ijbs.83368. eCollection 2023. Int J Biol Sci. 2023. PMID: 37416784 Free PMC article. Review.
-
Function of serine/arginine-rich splicing factors in hematopoiesis and hematopoietic malignancies.Cancer Cell Int. 2024 Jul 21;24(1):257. doi: 10.1186/s12935-024-03438-7. Cancer Cell Int. 2024. PMID: 39034387 Free PMC article. Review.
-
TEP RNA: a new frontier for early diagnosis of NSCLC.J Cancer Res Clin Oncol. 2024 Feb 19;150(2):97. doi: 10.1007/s00432-024-05620-w. J Cancer Res Clin Oncol. 2024. PMID: 38372784 Free PMC article. Review.
References
-
- Italiano JE Jr. Unraveling mechanisms that control platelet production. Semin Thromb Hemost. 2013;39(1):15-24. - PubMed
-
- Bluteau D, Lordier L, Di Stefano A, et al. . Regulation of megakaryocyte maturation and platelet formation. J Thromb Haemost. 2009;7(Suppl 1):227-234. - PubMed
-
- Nurden AT, Nurden P. Inherited disorders of platelet function: selected updates. J Thromb Haemost. 2015;13(Suppl 1):S2-S9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

