Cell cycle regulation of ER membrane biogenesis protects against chromosome missegregation
- PMID: 34852214
- PMCID: PMC8692360
- DOI: 10.1016/j.devcel.2021.11.009
Cell cycle regulation of ER membrane biogenesis protects against chromosome missegregation
Abstract
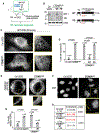
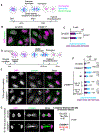
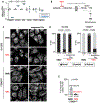
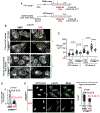
Failure to reorganize the endoplasmic reticulum (ER) in mitosis results in chromosome missegregation. Here, we show that accurate chromosome segregation in human cells requires cell cycle-regulated ER membrane production. Excess ER membranes increase the viscosity of the mitotic cytoplasm to physically restrict chromosome movements, which impedes the correction of mitotic errors leading to the formation of micronuclei. Mechanistically, we demonstrate that the protein phosphatase CTDNEP1 counteracts mTOR kinase to establish a dephosphorylated pool of the phosphatidic acid phosphatase lipin 1 in interphase. CTDNEP1 control of lipin 1 limits the synthesis of fatty acids for ER membrane biogenesis in interphase that then protects against chromosome missegregation in mitosis. Thus, regulation of ER size can dictate the biophysical properties of mitotic cells, providing an explanation for why ER reorganization is necessary for mitotic fidelity. Our data further suggest that dysregulated lipid metabolism is a potential source of aneuploidy in cancer cells.
Keywords: CTDNEP1; aneuploidy; lipid homeostasis; lipin; mTOR; medulloblastoma; micronuclei; mitosis; nuclear assembly.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Similar articles
-
Differential reliance of CTD-nuclear envelope phosphatase 1 on its regulatory subunit in ER lipid synthesis and storage.Mol Biol Cell. 2024 Jul 1;35(7):ar101. doi: 10.1091/mbc.E23-09-0382. Epub 2024 May 22. Mol Biol Cell. 2024. PMID: 38776127 Free PMC article.
-
Ice2 promotes ER membrane biogenesis in yeast by inhibiting the conserved lipin phosphatase complex.EMBO J. 2021 Nov 15;40(22):e107958. doi: 10.15252/embj.2021107958. Epub 2021 Oct 6. EMBO J. 2021. PMID: 34617598 Free PMC article.
-
Lipin-1 regulation of phospholipid synthesis maintains endoplasmic reticulum homeostasis and is critical for triple-negative breast cancer cell survival.FASEB J. 2017 Jul;31(7):2893-2904. doi: 10.1096/fj.201601353R. Epub 2017 Mar 27. FASEB J. 2017. PMID: 28347999 Free PMC article.
-
Lipin proteins and glycerolipid metabolism: Roles at the ER membrane and beyond.Biochim Biophys Acta Biomembr. 2017 Sep;1859(9 Pt B):1583-1595. doi: 10.1016/j.bbamem.2017.04.007. Epub 2017 Apr 11. Biochim Biophys Acta Biomembr. 2017. PMID: 28411173 Free PMC article. Review.
-
Mechanisms of nuclear envelope expansion.Curr Opin Cell Biol. 2024 Dec;91:102425. doi: 10.1016/j.ceb.2024.102425. Epub 2024 Sep 8. Curr Opin Cell Biol. 2024. PMID: 39250858 Review.
Cited by
-
Dynamics of CLIMP-63 S-acylation control ER morphology.Nat Commun. 2023 Jan 17;14(1):264. doi: 10.1038/s41467-023-35921-6. Nat Commun. 2023. PMID: 36650170 Free PMC article.
-
Research Trends in C-Terminal Domain Nuclear Envelope Phosphatase 1.Life (Basel). 2023 Jun 7;13(6):1338. doi: 10.3390/life13061338. Life (Basel). 2023. PMID: 37374122 Free PMC article. Review.
-
Loss of phosphatase CTDNEP1 potentiates aggressive medulloblastoma by triggering MYC amplification and genomic instability.Nat Commun. 2023 Feb 10;14(1):762. doi: 10.1038/s41467-023-36400-8. Nat Commun. 2023. PMID: 36765089 Free PMC article.
-
Baseline unfolded protein response signaling adjusts the timing of the mammalian cell cycle.Mol Biol Cell. 2024 Jun 1;35(6):br12. doi: 10.1091/mbc.E23-11-0419. Epub 2024 Apr 24. Mol Biol Cell. 2024. PMID: 38656789 Free PMC article.
-
A CRISPR-Cas9 screen reveals genetic determinants of the cellular response to decitabine.EMBO Rep. 2025 Mar;26(6):1528-1565. doi: 10.1038/s44319-025-00385-w. Epub 2025 Feb 10. EMBO Rep. 2025. PMID: 39930152 Free PMC article.
References
-
- Baumann O, and Walz B (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205, 149–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

