Synaptic protein interaction networks encode experience by assuming stimulus-specific and brain-region-specific states
- PMID: 34852231
- PMCID: PMC8722361
- DOI: 10.1016/j.celrep.2021.110076
Synaptic protein interaction networks encode experience by assuming stimulus-specific and brain-region-specific states
Abstract
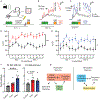
A core network of widely expressed proteins within the glutamatergic post-synapse mediates activity-dependent synaptic plasticity throughout the brain, but the specific proteomic composition of synapses differs between brain regions. Here, we address the question, how does proteomic composition affect activity-dependent protein-protein interaction networks (PINs) downstream of synaptic activity? Using quantitative multiplex co-immunoprecipitation, we compare the PIN response of in vivo or ex vivo neurons derived from different brain regions to activation by different agonists or different forms of eyeblink conditioning. We report that PINs discriminate between incoming stimuli using differential kinetics of overlapping and non-overlapping PIN parameters. Further, these "molecular logic rules" differ by brain region. We conclude that although the PIN of the glutamatergic post-synapse is expressed widely throughout the brain, its activity-dependent dynamics show remarkable stimulus-specific and brain-region-specific diversity. This diversity may help explain the challenges in developing molecule-specific drug therapies for neurological disorders.
Keywords: PiSCES; eyeblink conditioning; protein-protein interaction; signaling.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures

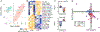




Similar articles
-
Long-lasting synaptic loss after repeated induction of LTD: independence to the means of LTD induction.Eur J Neurosci. 2006 Sep;24(6):1606-16. doi: 10.1111/j.1460-9568.2006.05032.x. Eur J Neurosci. 2006. PMID: 17004924
-
GPR30 activation improves memory and facilitates DHPG-induced LTD in the hippocampal CA3 of middle-aged mice.Neurobiol Learn Mem. 2018 Mar;149:10-19. doi: 10.1016/j.nlm.2018.02.005. Epub 2018 Feb 6. Neurobiol Learn Mem. 2018. PMID: 29421611
-
Long-term depression at olfactory nerve synapses.J Neurosci. 2005 Apr 27;25(17):4252-9. doi: 10.1523/JNEUROSCI.4721-04.2005. J Neurosci. 2005. PMID: 15858051 Free PMC article.
-
Rules of engagement: factors that regulate activity-dependent synaptic plasticity during neural network development.Biol Bull. 2010 Oct;219(2):81-99. doi: 10.1086/BBLv219n2p81. Biol Bull. 2010. PMID: 20972254 Review.
-
Activity-dependent development of GABAergic synapses.Brain Res. 2019 Mar 15;1707:18-26. doi: 10.1016/j.brainres.2018.11.014. Epub 2018 Nov 12. Brain Res. 2019. PMID: 30439352 Review.
Cited by
-
Differential protein-protein interactions underlie signaling mediated by the TCR and a 4-1BB domain-containing CAR.Sci Signal. 2024 Mar 5;17(826):eadd4671. doi: 10.1126/scisignal.add4671. Epub 2024 Mar 5. Sci Signal. 2024. PMID: 38442200 Free PMC article.
-
SRC family kinase inhibition rescues molecular and behavioral phenotypes, but not protein interaction network dynamics, in a mouse model of Fragile X syndrome.Mol Psychiatry. 2024 May;29(5):1392-1405. doi: 10.1038/s41380-024-02418-7. Epub 2024 Jan 31. Mol Psychiatry. 2024. PMID: 38297084 Free PMC article.
-
Integration across biophysical scales identifies molecular and cellular correlates of person-to-person variability in human brain connectivity.Nat Neurosci. 2024 Nov;27(11):2240-2252. doi: 10.1038/s41593-024-01788-z. Epub 2024 Oct 31. Nat Neurosci. 2024. PMID: 39482360 Free PMC article.
-
Protein interaction network analysis of mTOR signaling reveals modular organization.bioRxiv [Preprint]. 2023 Aug 4:2023.08.04.552011. doi: 10.1101/2023.08.04.552011. bioRxiv. 2023. Update in: J Biol Chem. 2023 Nov;299(11):105271. doi: 10.1016/j.jbc.2023.105271. PMID: 37577705 Free PMC article. Updated. Preprint.
-
Dissociation of the mTOR protein interaction network following neuronal activation is altered by Shank3 mutation.bioRxiv [Preprint]. 2025 May 23:2025.05.20.655155. doi: 10.1101/2025.05.20.655155. bioRxiv. 2025. PMID: 40475644 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

