A defective viral genome strategy elicits broad protective immunity against respiratory viruses
- PMID: 34852237
- PMCID: PMC8598942
- DOI: 10.1016/j.cell.2021.11.023
A defective viral genome strategy elicits broad protective immunity against respiratory viruses
Abstract
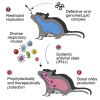
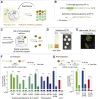
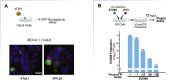
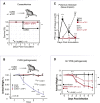
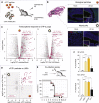
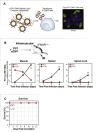
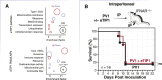
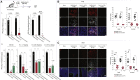
RNA viruses generate defective viral genomes (DVGs) that can interfere with replication of the parental wild-type virus. To examine their therapeutic potential, we created a DVG by deleting the capsid-coding region of poliovirus. Strikingly, intraperitoneal or intranasal administration of this genome, which we termed eTIP1, elicits an antiviral response, inhibits replication, and protects mice from several RNA viruses, including enteroviruses, influenza, and SARS-CoV-2. While eTIP1 replication following intranasal administration is limited to the nasal cavity, its antiviral action extends non-cell-autonomously to the lungs. eTIP1 broad-spectrum antiviral effects are mediated by both local and distal type I interferon responses. Importantly, while a single eTIP1 dose protects animals from SARS-CoV-2 infection, it also stimulates production of SARS-CoV-2 neutralizing antibodies that afford long-lasting protection from SARS-CoV-2 reinfection. Thus, eTIP1 is a safe and effective broad-spectrum antiviral generating short- and long-term protection against SARS-CoV-2 and other respiratory infections in animal models.
Keywords: RNA viruses; SARS-CoV-2; antiviral; broad-spectrum; defective viral genomes; innate immunity; interferon; respiratory infection.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Y.X., R.N., and R. Andino have submitted a patent application. Provisional patent application: recombinant enteroviruses and uses thereof. eTIP1. US Provisional Patent Filed 7/2020. The application was accorded serial no. 63/047,398. D.T. and R.N. are shareholders and employees of Aleph Therapeutics, Inc. E.B. is a shareholder and employee of Pine Biotech Inc.
Figures




Comment in
-
Defective viral genomes can protect against SARS-CoV-2 variants and other respiratory viruses.Nat Rev Immunol. 2022 Jan;22(1):5. doi: 10.1038/s41577-021-00668-y. Nat Rev Immunol. 2022. PMID: 34862485 Free PMC article. No abstract available.
-
Harnessing defective viruses to fight infections.Med. 2022 Jan 14;3(1):1-2. doi: 10.1016/j.medj.2021.12.006. Med. 2022. PMID: 35590140
References
-
- Ascough S., Vlachantoni I., Kalyan M., Haijema B.-J., Wallin-Weber S., Dijkstra-Tiekstra M., Ahmed M.S., van Roosmalen M., Grimaldi R., Zhang Q., et al. Local and Systemic Immunity against Respiratory Syncytial Virus Induced by a Novel Intranasal Vaccine. A Randomized, Double-Blind, Placebo-controlled Clinical Trial. Am. J. Respir. Crit. Care Med. 2019;200:481–492. - PMC - PubMed
-
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. - PubMed
-
- Bessière P., Wasniewski M., Picard-Meyer E., Servat A., Figueroa T., Foret-Lucas C., Coggon A., Lesellier S., Boué F., Cebron N., et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. PLoS Pathog. 2021;17:e1009427. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

