Receptor-binding domain-based immunoassays for serosurveillance differentiate efficiently between SARS-CoV2-exposed and non-exposed farmed mink
- PMID: 34852683
- PMCID: PMC8921823
- DOI: 10.1177/10406387211057859
Receptor-binding domain-based immunoassays for serosurveillance differentiate efficiently between SARS-CoV2-exposed and non-exposed farmed mink
Abstract
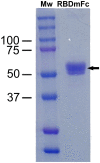
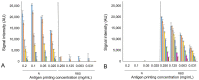
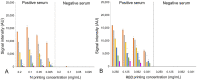
During the COVID-19 pandemic, infection of farmed mink has become not only an economic issue but also a widespread public health concern. International agencies have advised the use of strict molecular and serosurveillance methods for monitoring the SARS-CoV2 status on mink farms. We developed 2 ELISAs and a duplex protein microarray immunoassay (MI), all in a double-recognition format (DR), to detect SARS-CoV2 antibodies specific to the receptor-binding domain (RBD) of the spike protein and to the full-length nucleoprotein (N) in mink sera. We collected 264 mink serum samples and 126 oropharyngeal samples from 5 Spanish mink farms. In both of the ELISAs and the MI, RBD performed better than N protein for serologic differentiation of mink from SARS-CoV2-positive and -negative farms. Therefore, RBD was the optimal antigenic target for serosurveillance of mink farms.
Keywords: COVID-19; ELISA; RBD; SARS-CoV2; mink; protein microarray; serosurveillance.
Conflict of interest statement
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

