Using environmental DNA methods to improve detectability in an endangered sturgeon (Acipenser sinensis) monitoring program
- PMID: 34852759
- PMCID: PMC8638369
- DOI: 10.1186/s12862-021-01948-w
Using environmental DNA methods to improve detectability in an endangered sturgeon (Acipenser sinensis) monitoring program
Abstract
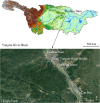
Background: To determine the presence and abundance of an aquatic species in large waterbodies, especially when populations are at low densities, is highly challenging for conservation biologists. Environmental DNA (eDNA) has the potential to offer a noninvasive and cost-effective method to complement traditional population monitoring, however, eDNA has not been extensively applied to study large migratory species. Chinese sturgeon (Acipenser sinensis), is the largest anadromous migratory fish in the Yangtze River, China, and in recent years its population has dramatically declined and spawning has failed, bringing this species to the brink of extinction. In this study, we aim to test the detectability of eDNA methods to determine the presence and relative abundance of reproductive stock of the species and whether eDNA can be used as a tool to reflect behavioral patterns. Chinese sturgeon eDNA was collected from four sites along the spawning ground across an eight month period, to investigate the temporal and spatial distribution using droplet digital PCR (ddPCR).
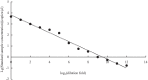
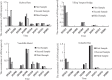
Results: We designed a pair of specific primers for Chinese sturgeon and demonstrated the high sensitivity of ddPCR to detect and quantify the Chinese sturgeon eDNA concentration with the limit of detection 0.17 copies/μl, with Chinese sturgeon eDNA been intermittently detected at all sampling sites. There was a consistent temporal pattern among four of the sampling sites that could reflect the movement characteristics of the Chinese sturgeon in the spawning ground, but without a spatial pattern. The eDNA concentration declined by approximately 2-3 × between December 2018 and December 2019.
Conclusions: The results prove the efficacy of eDNA for monitoring reproductive stock of the Chinese sturgeon and the e decreased eDNA concentration reflect that Chinese sturgeon may survive with an extremely small number of reproductive stock in the Yangtze River. Accordingly, we suggest future conservation measures should focus on both habitat restoration and matured fish restocking to ensure successful spawning. Overall, this study provides encouraging support for the application of eDNA methods to monitor endangered aquatic species.
Keywords: Droplet digital PCR; Environmental DNA; Reproductive stock; Sturgeon.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Monitoring seasonal distribution of an endangered anadromous sturgeon in a large river using environmental DNA.Naturwissenschaften. 2018 Oct 10;105(11-12):62. doi: 10.1007/s00114-018-1587-4. Naturwissenschaften. 2018. PMID: 30306348
-
Detection of Adult Green Sturgeon Using Environmental DNA Analysis.PLoS One. 2016 Apr 20;11(4):e0153500. doi: 10.1371/journal.pone.0153500. eCollection 2016. PLoS One. 2016. PMID: 27096433 Free PMC article.
-
Assessing the genetic diversity of the critically endangered Chinese sturgeon Acipenser sinensis using mitochondrial markers and genome-wide single-nucleotide polymorphisms from RAD-seq.Sci China Life Sci. 2018 Sep;61(9):1090-1098. doi: 10.1007/s11427-017-9254-6. Epub 2018 Mar 30. Sci China Life Sci. 2018. PMID: 29948902
-
Current status and topical issues on the use of eDNA-based targeted detection of rare animal species.Sci Total Environ. 2023 Dec 15;904:166675. doi: 10.1016/j.scitotenv.2023.166675. Epub 2023 Aug 28. Sci Total Environ. 2023. PMID: 37647964 Review.
-
Environmental DNA study on aquatic ecosystem monitoring and management: Recent advances and prospects.J Environ Manage. 2022 Dec 1;323:116310. doi: 10.1016/j.jenvman.2022.116310. Epub 2022 Sep 24. J Environ Manage. 2022. PMID: 36261997 Review.
Cited by
-
Correlation between the Density of Acipenser sinensis and Its Environmental DNA.Biology (Basel). 2023 Dec 28;13(1):19. doi: 10.3390/biology13010019. Biology (Basel). 2023. PMID: 38248450 Free PMC article.
References
-
- Thomsen PF, Willerslev E. Environmental DNA-an emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv. 2015;183:4–18.
-
- Shaw JL, Clarke LJ, Wedderburn SD, Barnes TC, Weyrich LS, Cooper A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol Conserv. 2016;197:131–138.
-
- Spear MJ, Embke HS, Krysan PJ, Vander Zanden MJ. Application of eDNA as a tool for assessing fish population abundance. Environ DNA. 2021;3:83–91.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
