Ensemble learning for the early prediction of neonatal jaundice with genetic features
- PMID: 34852805
- PMCID: PMC8638201
- DOI: 10.1186/s12911-021-01701-9
Ensemble learning for the early prediction of neonatal jaundice with genetic features
Abstract
Background: Neonatal jaundice may cause severe neurological damage if poorly evaluated and diagnosed when high bilirubin occurs. The study explored how to effectively integrate high-dimensional genetic features into predicting neonatal jaundice.
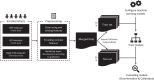
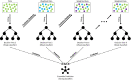
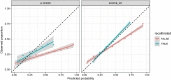
Methods: This study recruited 984 neonates from the Suzhou Municipal Central Hospital in China, and applied an ensemble learning approach to enhance the prediction of high-dimensional genetic features and clinical risk factors (CRF) for physiological neonatal jaundice of full-term newborns within 1-week after birth. Further, sigmoid recalibration was applied for validating the reliability of our methods.
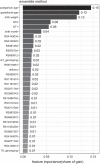
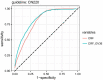
Results: The maximum accuracy of prediction reached 79.5% Area Under Curve (AUC) by CRF and could be marginally improved by 3.5% by including genetic variant (GV). Feature importance illustrated that 36 GVs contributed 55.5% in predicting neonatal jaundice in terms of gain from splits. Further analysis revealed that the main contribution of GV was to reduce the false-positive rate, i.e., to increase the specificity in the prediction.
Conclusions: Our study shed light on the theoretical and practical value of GV in the prediction of neonatal jaundice.
Keywords: Genetic variants; Hyperbilirubinemia; Machine learning; Transcutaneous bilirubin.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Bhutani VK, Stark AR, Lazzeroni LC, Poland R, Gourley GR, Kazmierczak S, Meloy L, Burgos AE, Hall JY, Stevenson DK, et al. Predischarge screening for severe neonatal hyperbilirubinemia identifies infants who need phototherapy. J Pediatr. 2013;162(3):477–482.e1. doi: 10.1016/j.jpeds.2012.08.022. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

