Observing the base-by-base search for native structure along transition paths during the folding of single nucleic acid hairpins
- PMID: 34853166
- PMCID: PMC8670488
- DOI: 10.1073/pnas.2101006118
Observing the base-by-base search for native structure along transition paths during the folding of single nucleic acid hairpins
Abstract
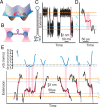
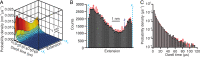
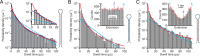
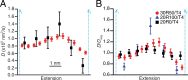
Biomolecular folding involves searching among myriad possibilities for the native conformation, but the elementary steps expected from theory for this search have never been detected directly. We probed the dynamics of folding at high resolution using optical tweezers, measuring individual trajectories as nucleic acid hairpins passed through the high-energy transition states that dominate kinetics and define folding mechanisms. We observed brief but ubiquitous pauses in the transition states, with a dwell time distribution that matched microscopic theories of folding quantitatively. The sequence dependence suggested that pauses were dominated by microbarriers from nonnative conformations during the search by each nucleotide residue for the native base-pairing conformation. Furthermore, the pauses were position dependent, revealing subtle local variations in energy-landscape roughness and allowing the diffusion coefficient describing the microscopic dynamics within the barrier to be found without reconstructing the shape of the energy landscape. These results show how high-resolution measurements can elucidate key microscopic events during folding to test fundamental theories of folding.
Keywords: diffusion; folding; kinetics; optical tweezers; transition states.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Bryngelson J. D., Wolynes P. G., Intermediates and barrier crossing in a random energy model (with applications to protein folding). J. Phys. Chem. 93, 6902–6915 (1989).
-
- Buchner J., Kiefhaber T., Protein Folding Handbook (Wiley-VCH, Weinheim, 2005).
-
- Hoffer N. Q., Woodside M. T., Probing microscopic conformational dynamics in folding reactions by measuring transition paths. Curr. Opin. Chem. Biol. 53, 68–74 (2019). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

