A point-mutation in the C-domain of CMP-sialic acid synthetase leads to lethality of medaka due to protein insolubility
- PMID: 34853329
- PMCID: PMC8636478
- DOI: 10.1038/s41598-021-01715-3
A point-mutation in the C-domain of CMP-sialic acid synthetase leads to lethality of medaka due to protein insolubility
Abstract
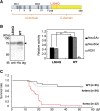
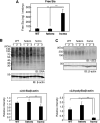
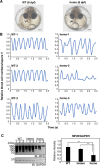
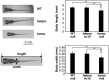
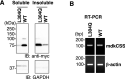
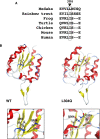
Vertebrate CMP-sialic acid synthetase (CSS), which catalyzes the synthesis of CMP-sialic acid (CMP-Sia), consists of a 28 kDa-N-domain and a 20 kDa-C-domain. The N-domain is known to be a catalytic domain; however, the significance of the C-domain still remains unknown. To elucidate the function of the C-domain at the organism level, we screened the medaka TILLING library and obtained medaka with non-synonymous mutations (t911a), or single amino acid substitutions of CSS, L304Q, in the C-domain. Prominently, most L304Q medaka was lethal within 19 days post-fertilization (dpf). L304Q young fry displayed free Sia accumulation, and impairment of sialylation, up to 8 dpf. At 8 dpf, a marked abnormality in ventricular contraction and skeletal myogenesis was observed. To gain insight into the mechanism of L304Q-induced abnormalities, L304Q was biochemically characterized. Although bacterially expressed soluble L304Q and WT showed the similar Vmax/Km values, very few soluble L304Q was detected when expressed in CHO cells in sharp contrast to the WT. Additionally, the thermostability of various mutations of L304 greatly decreased, except for WT and L304I. These results suggest that L304 is important for the stability of CSS, and that an appropriate level of expression of soluble CSS is significant for animal survival.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
A novel C-domain-dependent inhibition of the rainbow trout CMP-sialic acid synthetase activity by CMP-deaminoneuraminic acid.Biochem Biophys Res Commun. 2022 Aug 20;617(Pt 1):16-21. doi: 10.1016/j.bbrc.2022.05.031. Epub 2022 May 13. Biochem Biophys Res Commun. 2022. PMID: 35667241
-
Diverse subcellular localizations of the insect CMP-sialic acid synthetases.Glycobiology. 2017 Apr 1;27(4):329-341. doi: 10.1093/glycob/cww128. Glycobiology. 2017. PMID: 27986833
-
Identification of the nuclear export signals that regulate the intracellular localization of the mouse CMP-sialic acid synthetase.Biochem Biophys Res Commun. 2007 Mar 30;355(1):174-80. doi: 10.1016/j.bbrc.2007.01.139. Epub 2007 Feb 2. Biochem Biophys Res Commun. 2007. PMID: 17292865
-
CMP-Sialic Acid Synthetase: The Point of Constriction in the Sialylation Pathway.Top Curr Chem. 2015;366:139-67. doi: 10.1007/128_2013_477. Top Curr Chem. 2015. PMID: 24141690 Review.
-
CMP-sialic acid synthetase of the nucleus.Biochim Biophys Acta. 2004 Jul 6;1673(1-2):56-65. doi: 10.1016/j.bbagen.2004.04.006. Biochim Biophys Acta. 2004. PMID: 15238249 Review.
Cited by
-
Sulfation of sialic acid is ubiquitous and essential for vertebrate development.Sci Rep. 2022 Jul 21;12(1):12496. doi: 10.1038/s41598-022-15143-4. Sci Rep. 2022. PMID: 35864127 Free PMC article.
-
Nature-Inspired Intelligent Computing: A Comprehensive Survey.Research (Wash D C). 2024 Aug 16;7:0442. doi: 10.34133/research.0442. eCollection 2024. Research (Wash D C). 2024. PMID: 39156658 Free PMC article. Review.
-
CMP-sialic acid synthetase in Drosophila requires N-glycosylation of a noncanonical site.J Biol Chem. 2025 Jun;301(6):108483. doi: 10.1016/j.jbc.2025.108483. Epub 2025 Apr 7. J Biol Chem. 2025. PMID: 40204091 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

