SETD5 modulates homeostasis of hematopoietic stem cells by mediating RNA Polymerase II pausing in cooperation with HCF-1
- PMID: 34853439
- PMCID: PMC8979820
- DOI: 10.1038/s41375-021-01481-1
SETD5 modulates homeostasis of hematopoietic stem cells by mediating RNA Polymerase II pausing in cooperation with HCF-1
Abstract
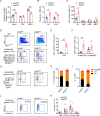
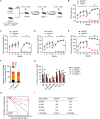
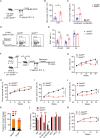
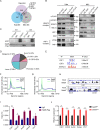
SETD5 mutations were identified as the genetic causes of neurodevelopmental disorders. While the whole-body knockout of Setd5 in mice leads to embryonic lethality, the role of SETD5 in adult stem cell remains unexplored. Here, a critical role of Setd5 in hematopoietic stem cells (HSCs) is identified. Specific deletion of Setd5 in hematopoietic system significantly increased the number of immunophenotypic HSCs by promoting HSC proliferation. Setd5-deficient HSCs exhibited impaired long-term self-renewal capacity and multiple-lineage differentiation potentials under transplantation pressure. Transcriptome analysis of Setd5-deficient HSCs revealed a disruption of quiescence state of long-term HSCs, a cause of the exhaustion of functional HSCs. Mechanistically, SETD5 was shown to regulate HSC quiescence by mediating the release of promoter-proximal paused RNA polymerase II (Pol II) on E2F targets in cooperation with HCF-1 and PAF1 complex. Taken together, these findings reveal an essential role of SETD5 in regulating Pol II pausing-mediated maintenance of adult stem cells.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation.Development. 2016 Dec 15;143(24):4595-4607. doi: 10.1242/dev.141465. Epub 2016 Nov 18. Development. 2016. PMID: 27864380 Free PMC article.
-
RNA polymerase II pausing modulates hematopoietic stem cell emergence in zebrafish.Blood. 2016 Sep 29;128(13):1701-10. doi: 10.1182/blood-2016-02-697847. Epub 2016 Aug 12. Blood. 2016. PMID: 27520065 Free PMC article.
-
SETD5 regulates the OGT-catalyzed O-GlcNAcylation of RNA polymerase II, which is involved in the stemness of colorectal cancer cells.Sci Rep. 2023 Nov 14;13(1):19885. doi: 10.1038/s41598-023-46923-1. Sci Rep. 2023. PMID: 37963940 Free PMC article.
-
Mitochondrial calcium homeostasis in hematopoietic stem cell: Molecular regulation of quiescence, function, and differentiation.Int Rev Cell Mol Biol. 2021;362:111-140. doi: 10.1016/bs.ircmb.2021.05.003. Epub 2021 Jun 21. Int Rev Cell Mol Biol. 2021. PMID: 34253293 Review.
-
Molecular regulation of hematopoietic stem cell quiescence.Cell Mol Life Sci. 2022 Mar 31;79(4):218. doi: 10.1007/s00018-022-04200-w. Cell Mol Life Sci. 2022. PMID: 35357574 Free PMC article. Review.
Cited by
-
SETD5 facilitates stemness and represses ferroptosis via m6A-mediating PKM2 stabilization in non-small cell lung cancer.Oncogene. 2025 Aug;44(29):2474-2489. doi: 10.1038/s41388-025-03426-9. Epub 2025 Apr 30. Oncogene. 2025. PMID: 40307507
-
Knockdown of SETD5 inhibited glycolysis and tumor growth in gastric cancer cells by down-regulating Akt signaling pathway.Open Life Sci. 2023 Oct 24;18(1):20220697. doi: 10.1515/biol-2022-0697. eCollection 2023. Open Life Sci. 2023. PMID: 37941780 Free PMC article.
-
Structure, activity and function of the lysine methyltransferase SETD5.Front Endocrinol (Lausanne). 2023 Feb 17;14:1089527. doi: 10.3389/fendo.2023.1089527. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36875494 Free PMC article. Review.
-
High Expression of Microtubule-associated Protein TBCB Predicts Adverse Outcome and Immunosuppression in Acute Myeloid Leukemia.J Cancer. 2023 Jun 12;14(10):1707-1724. doi: 10.7150/jca.84215. eCollection 2023. J Cancer. 2023. PMID: 37476188 Free PMC article.
-
Questions concerning tyrosine kinase-inhibitor therapy and transplants in chronic phase chronic myeloid leukaemia.Leukemia. 2022 May;36(5):1227-1236. doi: 10.1038/s41375-022-01522-3. Epub 2022 Mar 25. Leukemia. 2022. PMID: 35338251 Free PMC article. Review.
References
-
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

