Accuracy mechanism of eukaryotic ribosome translocation
- PMID: 34853469
- PMCID: PMC8674143
- DOI: 10.1038/s41586-021-04131-9
Accuracy mechanism of eukaryotic ribosome translocation
Abstract
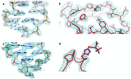
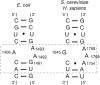
Translation of the genetic code into proteins is realized through repetitions of synchronous translocation of messenger RNA (mRNA) and transfer RNAs (tRNA) through the ribosome. In eukaryotes translocation is ensured by elongation factor 2 (eEF2), which catalyses the process and actively contributes to its accuracy1. Although numerous studies point to critical roles for both the conserved eukaryotic posttranslational modification diphthamide in eEF2 and tRNA modifications in supporting the accuracy of translocation, detailed molecular mechanisms describing their specific functions are poorly understood. Here we report a high-resolution X-ray structure of the eukaryotic 80S ribosome in a translocation-intermediate state containing mRNA, naturally modified eEF2 and tRNAs. The crystal structure reveals a network of stabilization of codon-anticodon interactions involving diphthamide1 and the hypermodified nucleoside wybutosine at position 37 of phenylalanine tRNA, which is also known to enhance translation accuracy2. The model demonstrates how the decoding centre releases a codon-anticodon duplex, allowing its movement on the ribosome, and emphasizes the function of eEF2 as a 'pawl' defining the directionality of translocation3. This model suggests how eukaryote-specific elements of the 80S ribosome, eEF2 and tRNAs undergo large-scale molecular reorganizations to ensure maintenance of the mRNA reading frame during the complex process of translocation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures








Similar articles
-
mRNA reading frame maintenance during eukaryotic ribosome translocation.Nature. 2024 Jan;625(7994):393-400. doi: 10.1038/s41586-023-06780-4. Epub 2023 Nov 29. Nature. 2024. PMID: 38030725
-
Atomic insights reveal fidelity mechanisms of eukaryotic protein synthesis.C R Biol. 2025 Jul 22;348:149-157. doi: 10.5802/crbiol.180. C R Biol. 2025. PMID: 40694004
-
Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.J Mol Biol. 2018 Aug 17;430(17):2677-2687. doi: 10.1016/j.jmb.2018.06.006. Epub 2018 Jun 7. J Mol Biol. 2018. PMID: 29886014
-
The ribosome's response to codon-anticodon mismatches.Biochimie. 2006 Aug;88(8):1001-11. doi: 10.1016/j.biochi.2006.04.013. Epub 2006 May 12. Biochimie. 2006. PMID: 16716484 Review.
-
Interactions of the ribosome with mRNA and tRNA.Curr Opin Struct Biol. 2010 Jun;20(3):325-32. doi: 10.1016/j.sbi.2010.03.002. Epub 2010 Apr 12. Curr Opin Struct Biol. 2010. PMID: 20392630 Review.
Cited by
-
Tuning tRNAs for improved translation.Front Genet. 2024 Jun 25;15:1436860. doi: 10.3389/fgene.2024.1436860. eCollection 2024. Front Genet. 2024. PMID: 38983271 Free PMC article. Review.
-
Structural basis of translation inhibition by a valine tRNA-derived fragment.Life Sci Alliance. 2024 Apr 10;7(6):e202302488. doi: 10.26508/lsa.202302488. Print 2024 Jun. Life Sci Alliance. 2024. PMID: 38599770 Free PMC article.
-
Capturing eukaryotic ribosome dynamics in situ at high resolution.Nat Struct Mol Biol. 2025 Apr;32(4):698-708. doi: 10.1038/s41594-024-01454-9. Epub 2025 Jan 9. Nat Struct Mol Biol. 2025. PMID: 39789210
-
Inconsistencies in the published rabbit ribosomal rRNAs: a proposal for uniformity in sequence and site numbering.RNA. 2025 May 16;31(6):781-790. doi: 10.1261/rna.080294.124. RNA. 2025. PMID: 40050069 Free PMC article.
-
Ribosome biogenesis: A central player in liver diseases.Genes Dis. 2025 Jan 4;12(5):101512. doi: 10.1016/j.gendis.2025.101512. eCollection 2025 Sep. Genes Dis. 2025. PMID: 40521004 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

