Human Cerebral Organoid Implantation Alleviated the Neurological Deficits of Traumatic Brain Injury in Mice
- PMID: 34853630
- PMCID: PMC8629662
- DOI: 10.1155/2021/6338722
Human Cerebral Organoid Implantation Alleviated the Neurological Deficits of Traumatic Brain Injury in Mice
Abstract
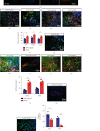
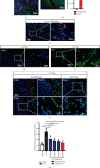
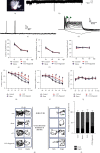
Traumatic brain injury (TBI) causes a high rate of mortality and disability, and its treatment is still limited. Loss of neurons in damaged area is hardly rescued by relative molecular therapies. Based on its disease characteristics, we transplanted human embryonic stem cell- (hESC-) derived cerebral organoids in the brain lesions of controlled cortical impact- (CCI-) modeled severe combined immunodeficient (SCID) mice. Grafted organoids survived and differentiated in CCI-induced lesion pools in mouse cortical tissue. Implanted cerebral organoids differentiated into various types of neuronal cells, extended long projections, and showed spontaneous action, as indicated by electromyographic activity in the grafts. Induced vascularization and reduced glial scar were also found after organoid implantation, suggesting grafting could improve local situation and promote neural repair. More importantly, the CCI mice's spatial learning and memory improved after organoid grafting. These findings suggest that cerebral organoid implanted in lesion sites differentiates into cortical neurons, forms long projections, and reverses deficits in spatial learning and memory, a potential therapeutic avenue for TBI.
Copyright © 2021 Zhongyuan Bao et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures



Similar articles
-
Functional recovery through the plastic adaptation of organoid grafts in cortical tissue.Cell Mol Life Sci. 2025 Jun 9;82(1):227. doi: 10.1007/s00018-025-05767-w. Cell Mol Life Sci. 2025. PMID: 40490613 Free PMC article. Review.
-
Therapeutic Transplantation of Human Central Nervous System Organoids for Neural Reconstruction.Int J Mol Sci. 2024 Aug 5;25(15):8540. doi: 10.3390/ijms25158540. Int J Mol Sci. 2024. PMID: 39126108 Free PMC article. Review.
-
Axonal Extensions along Corticospinal Tracts from Transplanted Human Cerebral Organoids.Stem Cell Reports. 2020 Aug 11;15(2):467-481. doi: 10.1016/j.stemcr.2020.06.016. Epub 2020 Jul 16. Stem Cell Reports. 2020. PMID: 32679062 Free PMC article.
-
Vascularization and Engraftment of Transplanted Human Cerebral Organoids in Mouse Cortex.eNeuro. 2018 Nov 20;5(6):ENEURO.0219-18.2018. doi: 10.1523/ENEURO.0219-18.2018. eCollection 2018 Nov-Dec. eNeuro. 2018. PMID: 30460331 Free PMC article.
-
Human cerebral organoids establish subcortical projections in the mouse brain after transplantation.Mol Psychiatry. 2021 Jul;26(7):2964-2976. doi: 10.1038/s41380-020-00910-4. Epub 2020 Oct 13. Mol Psychiatry. 2021. PMID: 33051604 Free PMC article.
Cited by
-
Organoids and chimeras: the hopeful fusion transforming traumatic brain injury research.Acta Neuropathol Commun. 2024 Aug 30;12(1):141. doi: 10.1186/s40478-024-01845-5. Acta Neuropathol Commun. 2024. PMID: 39215375 Free PMC article. Review.
-
Murine Xenograft Models as Preclinical Tools in Endometrial Cancer Research.Cancers (Basel). 2024 Nov 28;16(23):3994. doi: 10.3390/cancers16233994. Cancers (Basel). 2024. PMID: 39682182 Free PMC article. Review.
-
Stem Cell Therapy in Neonatal Hypoxic-Ischemic Encephalopathy and Cerebral Palsy: a Bibliometric Analysis and New Strategy.Mol Neurobiol. 2024 Jul;61(7):4538-4564. doi: 10.1007/s12035-023-03848-0. Epub 2023 Dec 16. Mol Neurobiol. 2024. PMID: 38102517 Review.
-
Functional recovery through the plastic adaptation of organoid grafts in cortical tissue.Cell Mol Life Sci. 2025 Jun 9;82(1):227. doi: 10.1007/s00018-025-05767-w. Cell Mol Life Sci. 2025. PMID: 40490613 Free PMC article. Review.
-
Therapeutic Transplantation of Human Central Nervous System Organoids for Neural Reconstruction.Int J Mol Sci. 2024 Aug 5;25(15):8540. doi: 10.3390/ijms25158540. Int J Mol Sci. 2024. PMID: 39126108 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

