Melanoma susceptibility: an update on genetic and epigenetic findings
- PMID: 34853632
- PMCID: PMC8611230
Melanoma susceptibility: an update on genetic and epigenetic findings
Abstract
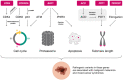
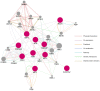
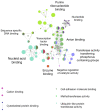
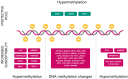
Malignant melanoma is one of the most highly ranked cancers in terms of years of life lost. Hereditary melanoma with its increased familial susceptibility is thought to affect up to 12% of all melanoma patients. In the past, only a few high-penetrance genes associated with familial melanoma, such as CDKN2A and CDK4, have been clinically tested. However, findings now indicate that melanoma is a cancer most likely to develop not only due to high-penetrance variants but also due to polygenic inheritance patterns, leaving no clear division between the hereditary and sporadic development of malignant melanoma. Various pathogenic low-penetrance variants were recently discovered through genome-wide association studies, and are now translated into polygenic risk scores. These can show superior sensitivity rates for the prediction of melanoma susceptibility and related mixed cancer syndromes than risk scores based on phenotypic traits of the patients, with odds ratios of up to 5.7 for patients in risk groups. In addition to describing genetic findings, we also review the first results of epigenetic research showing constitutional methylation changes that alter the susceptibility to cutaneous melanoma and its risk factors.
Keywords: Melanoma susceptibility; epigenetics; familial melanoma; germline mutation; hereditary melanoma; melanoma genetics; polygenic risk score.
IJMEG Copyright © 2021.
Conflict of interest statement
None.
Figures
Similar articles
-
Multiplex melanoma families are enriched for polygenic risk.Hum Mol Genet. 2020 Oct 10;29(17):2976-2985. doi: 10.1093/hmg/ddaa156. Hum Mol Genet. 2020. PMID: 32716505 Free PMC article.
-
Melanoma genetics.J Med Genet. 2016 Jan;53(1):1-14. doi: 10.1136/jmedgenet-2015-103150. Epub 2015 Sep 3. J Med Genet. 2016. PMID: 26337759 Review.
-
Familial Melanoma and Susceptibility Genes: A Review of the Most Common Clinical and Dermoscopic Phenotypic Aspect, Associated Malignancies and Practical Tips for Management.J Clin Med. 2021 Aug 23;10(16):3760. doi: 10.3390/jcm10163760. J Clin Med. 2021. PMID: 34442055 Free PMC article. Review.
-
Absence of germline CDKN2A mutation in Sicilian patients with familial malignant melanoma: Could it be a population-specific genetic signature?Cancer Biol Ther. 2016;17(1):83-90. doi: 10.1080/15384047.2015.1108494. Cancer Biol Ther. 2016. PMID: 26650572 Free PMC article.
-
CDKN2A and CDK4 variants in Latvian melanoma patients: analysis of a clinic-based population.Melanoma Res. 2007 Jun;17(3):185-91. doi: 10.1097/CMR.0b013e328014a2cd. Melanoma Res. 2007. PMID: 17505264
Cited by
-
Malignant Melanoma in a Retrospective Cohort of Immunocompromised Patients: A Statistical and Pathologic Analysis.Cancers (Basel). 2023 Jul 13;15(14):3600. doi: 10.3390/cancers15143600. Cancers (Basel). 2023. PMID: 37509262 Free PMC article.
-
Prevalence of CDKN2A, CDK4, POT1, BAP1, MITF, ATM, and TERT Pathogenic Variants in a Single-Center Retrospective Series of Patients With Melanoma and Personal or Family History Suggestive of Genetic Predisposition.Dermatol Pract Concept. 2024 Jul 1;14(3):e2024120. doi: 10.5826/dpc.1403a120. Dermatol Pract Concept. 2024. PMID: 39122510 Free PMC article.
-
Melanoma risk prediction based on a polygenic risk score and clinical risk factors.Melanoma Res. 2023 Aug 1;33(4):293-299. doi: 10.1097/CMR.0000000000000896. Epub 2023 Apr 24. Melanoma Res. 2023. PMID: 37096571 Free PMC article.
-
Epigenetic Modification of PD-1/PD-L1-Mediated Cancer Immunotherapy against Melanoma.Int J Mol Sci. 2022 Jan 20;23(3):1119. doi: 10.3390/ijms23031119. Int J Mol Sci. 2022. PMID: 35163049 Free PMC article. Review.
-
Ruthenium(II) Complex with 1-Hydroxy-9,10-Anthraquinone Inhibits Cell Cycle Progression at G0/G1 and Induces Apoptosis in Melanoma Cells.Pharmaceuticals (Basel). 2025 Jan 8;18(1):63. doi: 10.3390/ph18010063. Pharmaceuticals (Basel). 2025. PMID: 39861126 Free PMC article.
References
-
- Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–1011. - PubMed
-
- Shimizu H. (Malignant) melanoma. In: Shimizu H, editor. Shimizu’s dermatology. 2nd edition. Chichester: John Wiley & Sons, Ltd.; 2017. pp. 521–527.
-
- Guy GP, Ekwueme DU. Years of potential life lost and indirect costs of melanoma and non-melanoma skin cancer: a systematic review of the literature. Pharmacoeconomics. 2011;29:863–874. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
