Clinical Assessment and Validation of a Rapid and Sensitive SARS-CoV-2 Test Using Reverse Transcription Loop-Mediated Isothermal Amplification Without the Need for RNA Extraction
- PMID: 34853795
- PMCID: PMC7798487
- DOI: 10.1093/ofid/ofaa631
Clinical Assessment and Validation of a Rapid and Sensitive SARS-CoV-2 Test Using Reverse Transcription Loop-Mediated Isothermal Amplification Without the Need for RNA Extraction
Abstract
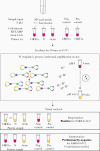
Background: Amid the enduring pandemic, there is an urgent need for expanded access to rapid, sensitive, and inexpensive coronavirus disease 2019 (COVID-19) testing worldwide without specialized equipment. We developed a simple test that uses colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) to detect severe acute resrpiratory syndrome coronavirus 2 (SARS-CoV-2) in 40 minutes from sample collection to result.
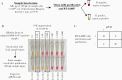
Methods: We tested 135 nasopharyngeal specimens from patients evaluated for COVID-19 infection at Massachusetts General Hospital. Specimens were either added directly to RT-LAMP reactions, inactivated by a combined chemical and heat treatment step, or inactivated then purified with a silica particle-based concentration method. Amplification was performed with 2 SARS-CoV-2-specific primer sets and an internal specimen control; the resulting color change was visually interpreted.
Results: Direct RT-LAMP testing of unprocessed specimens could only reliably detect samples with abundant SARS-CoV-2 (>3 000 000 copies/mL), with sensitivities of 50% (95% CI, 28%-72%) and 59% (95% CI, 43%-73%) in samples collected in universal transport medium and saline, respectively, compared with quantitative polymerase chain reaction (qPCR). Adding an upfront RNase inactivation step markedly improved the limit of detection to at least 25 000 copies/mL, with 87.5% (95% CI, 72%-95%) sensitivity and 100% specificity (95% CI, 87%-100%). Using both inactivation and purification increased the assay sensitivity by 10-fold, achieving a limit of detection comparable to commercial real-time PCR-based diagnostics.
Conclusions: By incorporating a fast and inexpensive sample preparation step, RT-LAMP accurately detects SARS-CoV-2 with limited equipment for about US$6 per sample, making this a potentially ideal assay to increase testing capacity, especially in resource-limited settings.
Keywords: COVID-19; LAMP; SARS-CoV-2; diagnostics; isothermal amplification; nucleic acid technology; rapid tests.
© The Author(s) 2020. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Figures


References
-
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 2002; 16:223–9. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

