Characterization of a library of 20 HBV-specific MHC class II-restricted T cell receptors
- PMID: 34853796
- PMCID: PMC8605085
- DOI: 10.1016/j.omtm.2021.10.012
Characterization of a library of 20 HBV-specific MHC class II-restricted T cell receptors
Abstract
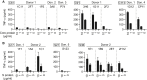
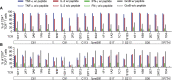
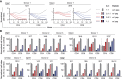
CD4+ T cells play an important role in the immune response against cancer and infectious diseases. However, mechanistic details of their helper function in hepatitis B virus (HBV) infection in particular, or their advantage for adoptive T cell therapy remain poorly understood as experimental and therapeutic tools are missing. Therefore, we identified, cloned, and characterized a comprehensive library of 20 MHC class II-restricted HBV-specific T cell receptors (TCRs) from donors with acute or resolved HBV infection. The TCRs were restricted by nine different MHC II molecules and specific for eight different epitopes derived from intracellularly processed HBV envelope, core, and polymerase proteins. Retroviral transduction resulted in a robust expression of all TCRs on primary T cells. A high functional avidity was measured for all TCRs specific for epitopes S17, S21, S36, and P774 (half-maximal effective concentration [EC50] <10 nM), or C61 and preS9 (EC50 <100 nM). Eight TCRs recognized peptide variants of HBV genotypes A to D. Both CD4+ and CD8+ T cells transduced with the MHC II-restricted TCRs were polyfunctional, producing interferon (IFN)-γ, tumor necrosis factor (TNF)-α, interleukin (IL)-2, and granzyme B (GrzB), and killed peptide-loaded target cells. Our set of MHC class II-restricted TCRs represents an important tool for elucidating CD4+ T cell help in viral infection with potential benefit for T cell therapy.
Keywords: CD4+ T cells; HBV clearance; MHC class II-restricted T cell receptors; T cell help; TCR expression; adoptive T cell therapy; chronic hepatitis B; hepatitis B virus infection; hepatocellular carcinoma; retroviral transduction.
© 2021 The Authors.
Conflict of interest statement
K. Wisskirchen is partially employed by SCG Cell Therapy GmbH and holds shares of SCG Cell Therapy Pte. Ltd. U.P. holds shares and received research funding from SCG Cell Therapy Pte. Ltd. The other authors declare no competing interests.
Figures





Similar articles
-
Isolation and functional characterization of hepatitis B virus-specific T-cell receptors as new tools for experimental and clinical use.PLoS One. 2017 Aug 8;12(8):e0182936. doi: 10.1371/journal.pone.0182936. eCollection 2017. PLoS One. 2017. PMID: 28792537 Free PMC article.
-
TNF-α/IFN-γ profile of HBV-specific CD4 T cells is associated with liver damage and viral clearance in chronic HBV infection.J Hepatol. 2020 Jan;72(1):45-56. doi: 10.1016/j.jhep.2019.08.024. Epub 2019 Sep 6. J Hepatol. 2020. PMID: 31499130
-
T cell recognition of hepatitis B and C viral antigens.Eur J Clin Invest. 1994 Oct;24(10):641-50. doi: 10.1111/j.1365-2362.1994.tb01055.x. Eur J Clin Invest. 1994. PMID: 7531642 Review.
-
OX40 stimulation and PD-L1 blockade synergistically augment HBV-specific CD4 T cells in patients with HBeAg-negative infection.J Hepatol. 2019 Jun;70(6):1103-1113. doi: 10.1016/j.jhep.2019.02.016. Epub 2019 Feb 28. J Hepatol. 2019. PMID: 30826436
-
Functional Signatures of Human CD4 and CD8 T Cell Responses to Mycobacterium tuberculosis.Front Immunol. 2014 Apr 22;5:180. doi: 10.3389/fimmu.2014.00180. eCollection 2014. Front Immunol. 2014. PMID: 24795723 Free PMC article. Review.
Cited by
-
Host and HBV Interactions and Their Potential Impact on Clinical Outcomes.Pathogens. 2023 Sep 8;12(9):1146. doi: 10.3390/pathogens12091146. Pathogens. 2023. PMID: 37764954 Free PMC article. Review.
-
Engineering HBV-specific T cells for the treatment of HBV-related HCC and HBV infection: Past, Present, and Future. Editorial on "Genetically-modified, redirected T cells target hepatitis B surface antigen-positive hepatocytes and hepatocellular carcinoma lesions in a clinical setting".Clin Mol Hepatol. 2024 Oct;30(4):728-734. doi: 10.3350/cmh.2024.0469. Epub 2024 Jun 27. Clin Mol Hepatol. 2024. PMID: 38934109 Free PMC article. No abstract available.
-
Exploiting TCR Repertoire Analysis to Select Therapeutic TCRs for Cancer Immunotherapy.Cells. 2025 Aug 7;14(15):1223. doi: 10.3390/cells14151223. Cells. 2025. PMID: 40801654 Free PMC article. Review.
-
The HLA class-II immunopeptidomes of AAV capsids proteins.Front Immunol. 2022 Dec 20;13:1067399. doi: 10.3389/fimmu.2022.1067399. eCollection 2022. Front Immunol. 2022. PMID: 36605211 Free PMC article.
-
Activation of distinct antiviral T-cell immunity: A comparison of bi- and trispecific T-cell engager antibodies with a chimeric antigen receptor targeting HBV envelope proteins.Front Immunol. 2022 Nov 3;13:1029214. doi: 10.3389/fimmu.2022.1029214. eCollection 2022. Front Immunol. 2022. PMID: 36405686 Free PMC article.
References
-
- WHO Hepatitis B, Fact Sheet. 2021 https://www.who.int/news-room/fact-sheets/detail/hepatitis-b
-
- Bohne F., Protzer U. Adoptive T-cell therapy as a therapeutic option for chronic hepatitis B. J. Viral Hepat. 2007;14(Suppl 1):45–50. - PubMed
-
- Ilan Y., Nagler A., Adler R., Naparstek E., Or R., Slavin S., Brautbar C., Shouval D. Adoptive transfer of immunity to hepatitis B virus after T cell-depleted allogeneic bone marrow transplantation. Hepatology. 1993;18:246–252. - PubMed
-
- Lau G.K., Lok A.S., Liang R.H., Lai C.L., Chiu E.K., Lau Y.L., Lam S.K. Clearance of hepatitis B surface antigen after bone marrow transplantation: role of adoptive immunity transfer. Hepatology. 1997;25:1497–1501. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

