Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: The proteomics-guided metabolic engineering
- PMID: 34853817
- PMCID: PMC8605246
- DOI: 10.1016/j.synbio.2021.11.003
Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: The proteomics-guided metabolic engineering
Abstract
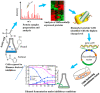
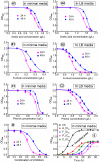
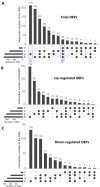
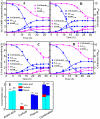
Pretreatment of lignocellulosic biomass is crucial for the release of biofermentable sugars for biofuels production, which could greatly alleviate the burgeoning environment and energy crisis caused by the massive usage of traditional fossil fuels. Pyrolysis is a cost-saving pretreatment process that can readily decompose biomass into levoglucosan, a promising anhydrosugar; however, many undesired toxic compounds inhibitory to downstream microbial fermentation are also generated during the pyrolysis, immensely impeding the bioconversion of levoglucosan-containing pyrolysate. Here, we took the first insight into the proteomic responses of a levoglucosan-utilizing and ethanol-producing Escherichia coli to three representative biomass-derived inhibitors, identifying large amounts of differentially expressed proteins (DEPs) that could guide the downstream metabolic engineering for the development of inhibitor-resistant strains. Fifteen up- and eight down-regulated DEPs were further identified as the biomarker stress-responsive proteins candidate for cellular tolerance to multiple inhibitors. Among these biomarker proteins, YcfR exhibiting the highest expression fold-change level was chosen as the target of overexpression to validate proteomics results and develop robust strains with enhanced inhibitor tolerance and fermentation performance. Finally, based on four plasmid-borne genes encoding the levoglucosan kinase, pyruvate decarboxylase, alcohol dehydrogenase, and protein YcfR, a new recombinant strain E. coli LGE-ycfR was successfully created, showing much higher acetic acid-, furfural-, and phenol-tolerance levels compared to the control without overexpression of ycfR. The specific growth rate, final cell density, ethanol concentration, ethanol productivity, and levoglucosan consumption rate of the recombinant were also remarkably improved. From the proteomics-guided metabolic engineering and phenotypic observations, we for the first time corroborated that YcfR is a stress-induced protein responsive to multiple biomass-derived inhibitors, and also developed an inhibitors-resistant strain that could produce bioethanol from levoglucosan in the presence of inhibitors of relatively high concentration. The newly developed E. coli LGE-ycfR strain that could eliminate the commonly-used costly detoxicification processes, is of great potential for the in situ cost-effective bioethanol production from the biomass-derived pyrolytic substrates.
Keywords: Bioethanol; Gene overexpression; Inhibitor; Levoglucosan; Lignocellulosic biomass; Proteomics.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work of this paper.
Figures


Similar articles
-
Mutations in adaptively evolved Escherichia coli LGE2 facilitated the cost-effective upgrading of undetoxified bio-oil to bioethanol fuel.Bioresour Bioprocess. 2021 Oct 22;8(1):105. doi: 10.1186/s40643-021-00459-2. Bioresour Bioprocess. 2021. PMID: 38650237 Free PMC article.
-
Proteomic and metabolomic analysis of the cellular biomarkers related to inhibitors tolerance in Zymomonas mobilis ZM4.Biotechnol Biofuels. 2018 Oct 16;11:283. doi: 10.1186/s13068-018-1287-5. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30356850 Free PMC article.
-
Omics analysis coupled with gene editing revealed potential transporters and regulators related to levoglucosan metabolism efficiency of the engineered Escherichia coli.Biotechnol Biofuels Bioprod. 2022 Jan 11;15(1):2. doi: 10.1186/s13068-022-02102-4. Biotechnol Biofuels Bioprod. 2022. PMID: 35418138 Free PMC article.
-
Microbial conversion of pyrolytic products to biofuels: a novel and sustainable approach toward second-generation biofuels.J Ind Microbiol Biotechnol. 2015 Dec;42(12):1557-79. doi: 10.1007/s10295-015-1687-5. Epub 2015 Oct 3. J Ind Microbiol Biotechnol. 2015. PMID: 26433384 Review.
-
Engineering Ligninolytic Consortium for Bioconversion of Lignocelluloses to Ethanol and Chemicals.Protein Pept Lett. 2018;25(2):108-119. doi: 10.2174/0929866525666180122105835. Protein Pept Lett. 2018. PMID: 29359652 Review.
Cited by
-
Microbial detoxification of lignocellulosic biomass hydrolysates: Biochemical and molecular aspects, challenges, exploits and future perspectives.Front Bioeng Biotechnol. 2022 Nov 22;10:1061667. doi: 10.3389/fbioe.2022.1061667. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36483774 Free PMC article. Review.
-
Kinase expression enhances phenolic aldehydes conversion and ethanol fermentability of Zymomonas mobilis.Bioprocess Biosyst Eng. 2022 Aug;45(8):1319-1329. doi: 10.1007/s00449-022-02747-3. Epub 2022 Jul 3. Bioprocess Biosyst Eng. 2022. PMID: 35786774
References
-
- Rubin E.M. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. - PubMed
-
- Anex R.P., Aden A., Kazi F.K., Fortman J., Swanson R.M., Wright M.M., Satrio J.A., Brown R.C., Daugaard D.E., Platon A. Techno-economic comparison of biomass-to-transportation fuels via pyrolysis, gasification, and biochemical pathways. Fuel. 2010;89:S29–S35.
-
- Islam Z.U., Zhisheng Y., Hassan el B., Dongdong C., Hongxun Z. Microbial conversion of pyrolytic products to biofuels: a novel and sustainable approach toward second-generation biofuels. J Ind Microbiol Biotechnol. 2015;42:1557–1579. - PubMed
LinkOut - more resources
Full Text Sources

