Live imaging of transcription sites using an elongating RNA polymerase II-specific probe
- PMID: 34854870
- PMCID: PMC8647360
- DOI: 10.1083/jcb.202104134
Live imaging of transcription sites using an elongating RNA polymerase II-specific probe
Abstract
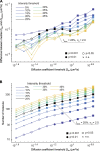
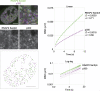
In eukaryotic nuclei, most genes are transcribed by RNA polymerase II (RNAP2), whose regulation is a key to understanding the genome and cell function. RNAP2 has a long heptapeptide repeat (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7), and Ser2 is phosphorylated on an elongation form. To detect RNAP2 Ser2 phosphorylation (RNAP2 Ser2ph) in living cells, we developed a genetically encoded modification-specific intracellular antibody (mintbody) probe. The RNAP2 Ser2ph-mintbody exhibited numerous foci, possibly representing transcription "factories," and foci were diminished during mitosis and in a Ser2 kinase inhibitor. An in vitro binding assay using phosphopeptides confirmed the mintbody's specificity. RNAP2 Ser2ph-mintbody foci were colocalized with proteins associated with elongating RNAP2 compared with factors involved in the initiation. These results support the view that mintbody localization represents the sites of RNAP2 Ser2ph in living cells. RNAP2 Ser2ph-mintbody foci showed constrained diffusional motion like chromatin, but they were more mobile than DNA replication domains and p300-enriched foci, suggesting that the elongating RNAP2 complexes are separated from more confined chromatin domains.
© 2021 Uchino et al.
Figures









Similar articles
-
Organization and Dynamics of Transcription Elongation Foci in Mouse Tissues.J Mol Biol. 2025 Aug 13:169395. doi: 10.1016/j.jmb.2025.169395. Online ahead of print. J Mol Biol. 2025. PMID: 40816716
-
Imaging transcription elongation dynamics by new technologies unveils the organization of initiation and elongation in transcription factories.Curr Opin Cell Biol. 2022 Feb;74:71-79. doi: 10.1016/j.ceb.2022.01.002. Epub 2022 Feb 17. Curr Opin Cell Biol. 2022. PMID: 35183895 Review.
-
Histone H3K27 acetylation precedes active transcription during zebrafish zygotic genome activation as revealed by live-cell analysis.Development. 2019 Sep 30;146(19):dev179127. doi: 10.1242/dev.179127. Development. 2019. PMID: 31570370 Free PMC article.
-
Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation.EMBO J. 2012 Jun 13;31(12):2784-97. doi: 10.1038/emboj.2012.123. Epub 2012 May 1. EMBO J. 2012. PMID: 22549466 Free PMC article.
-
Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription.Biochim Biophys Acta. 2013 Jan;1829(1):55-62. doi: 10.1016/j.bbagrm.2012.08.013. Epub 2012 Sep 7. Biochim Biophys Acta. 2013. PMID: 22982363 Review.
Cited by
-
Characterizing Properties of Biomolecular Condensates Below the Diffraction Limit In Vivo.Methods Mol Biol. 2023;2563:425-445. doi: 10.1007/978-1-0716-2663-4_22. Methods Mol Biol. 2023. PMID: 36227487
-
Mode of SUV420H2 heterochromatin localization through multiple HP1 binding motifs in the heterochromatic targeting module.Genes Cells. 2024 May;29(5):361-379. doi: 10.1111/gtc.13109. Epub 2024 Feb 25. Genes Cells. 2024. PMID: 38403935 Free PMC article.
-
Feeling the force from within - new tools and insights into nuclear mechanotransduction.J Cell Sci. 2025 Mar 1;138(5):JCS263615. doi: 10.1242/jcs.263615. Epub 2025 Mar 10. J Cell Sci. 2025. PMID: 40059756 Review.
-
Live-cell imaging uncovers the relationship between histone acetylation, transcription initiation, and nucleosome mobility.Sci Adv. 2023 Oct 6;9(40):eadh4819. doi: 10.1126/sciadv.adh4819. Epub 2023 Oct 4. Sci Adv. 2023. PMID: 37792937 Free PMC article.
-
AI-assisted protein design to rapidly convert antibody sequences to intrabodies targeting diverse peptides and histone modifications.bioRxiv [Preprint]. 2025 Feb 9:2025.02.06.636921. doi: 10.1101/2025.02.06.636921. bioRxiv. 2025. PMID: 39975170 Free PMC article. Preprint.
References
-
- Boehning, M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P., et al. . 2018. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25:833–840. 10.1038/s41594-018-0112-y - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

