Quantitative subcellular acyl-CoA analysis reveals distinct nuclear metabolism and isoleucine-dependent histone propionylation
- PMID: 34856123
- PMCID: PMC8950487
- DOI: 10.1016/j.molcel.2021.11.006
Quantitative subcellular acyl-CoA analysis reveals distinct nuclear metabolism and isoleucine-dependent histone propionylation
Abstract
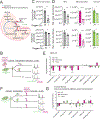
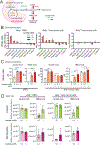
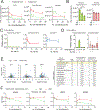
Quantitative subcellular metabolomic measurements can explain the roles of metabolites in cellular processes but are subject to multiple confounding factors. We developed stable isotope labeling of essential nutrients in cell culture-subcellular fractionation (SILEC-SF), which uses isotope-labeled internal standard controls that are present throughout fractionation and processing to quantify acyl-coenzyme A (acyl-CoA) thioesters in subcellular compartments by liquid chromatography-mass spectrometry. We tested SILEC-SF in a range of sample types and examined the compartmentalized responses to oxygen tension, cellular differentiation, and nutrient availability. Application of SILEC-SF to the challenging analysis of the nuclear compartment revealed a nuclear acyl-CoA profile distinct from that of the cytosol, with notable nuclear enrichment of propionyl-CoA. Using isotope tracing, we identified the branched chain amino acid isoleucine as a major metabolic source of nuclear propionyl-CoA and histone propionylation, thus revealing a new mechanism of crosstalk between metabolism and the epigenome.
Keywords: acyl-CoA; branched chain amino acids; histone; internal standard; isoleucine; matrix effects; metabolomics; mitochondria; nucleus; propionylation; subcellular.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



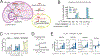
References
-
- Bedi KC, Snyder NW, Brandimarto J, Aziz M, Mesaros C, Worth AJ, Wang LL, Javaheri A, Blair IA, Margulies KB, et al. (2016). Evidence for Intramyocardial Disruption of Lipid Metabolism and Increased Myocardial Ketone Utilization in Advanced Human Heart Failure. Circulation 133, 706–716. - PMC - PubMed
-
- Boon R, Silveira GG, and Mostoslavsky R. (2020). Nuclear metabolism and the regulation of the epigenome. Nat. Metab 1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM132261/GM/NIGMS NIH HHS/United States
- K00 CA222741/CA/NCI NIH HHS/United States
- R01 CA174761/CA/NCI NIH HHS/United States
- S10 OD030286/OD/NIH HHS/United States
- T32 GM007229/GM/NIGMS NIH HHS/United States
- T32 GM007170/GM/NIGMS NIH HHS/United States
- R01 DK116005/DK/NIDDK NIH HHS/United States
- W 1226/FWF_/Austrian Science Fund FWF/Austria
- P30 ES013508/ES/NIEHS NIH HHS/United States
- R01 CA248315/CA/NCI NIH HHS/United States
- R01 CA228339/CA/NCI NIH HHS/United States
- T32 CA115299/CA/NCI NIH HHS/United States
- F99 CA222741/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

