Human blastoids model blastocyst development and implantation
- PMID: 34856602
- PMCID: PMC8791832
- DOI: 10.1038/s41586-021-04267-8
Human blastoids model blastocyst development and implantation
Abstract
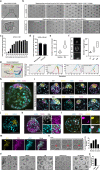
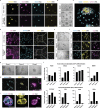
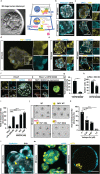
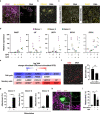
One week after fertilization, human embryos implant into the uterus. This event requires the embryo to form a blastocyst consisting of a sphere encircling a cavity lodging the embryo proper. Stem cells can form a blastocyst model that we called a blastoid1. Here we show that naive human pluripotent stem cells cultured in PXGL medium2 and triply inhibited for the Hippo, TGF-β and ERK pathways efficiently (with more than 70% efficiency) form blastoids generating blastocyst-stage analogues of the three founding lineages (more than 97% trophectoderm, epiblast and primitive endoderm) according to the sequence and timing of blastocyst development. Blastoids spontaneously form the first axis, and we observe that the epiblast induces the local maturation of the polar trophectoderm, thereby endowing blastoids with the capacity to directionally attach to hormonally stimulated endometrial cells, as during implantation. Thus, we propose that such a human blastoid is a faithful, scalable and ethical model for investigating human implantation and development3,4.
© 2021. The Author(s).
Conflict of interest statement
The Institute for Molecular Biotechnology, Austrian Academy of Sciences has filed patent application EP21151455.9 describing the protocols for human blastoid formation and for the blastoid–endometrium interaction assay. H.K., A.J., H.H.K. and N.R. are the inventors on this patent. All other authors declare no competing interests.
Figures









Comment in
-
A role model of human blastocysts.Nat Rev Mol Cell Biol. 2022 Feb;23(2):91. doi: 10.1038/s41580-021-00445-8. Nat Rev Mol Cell Biol. 2022. PMID: 34907363 No abstract available.
-
Human blastoids 3.0.Cell Stem Cell. 2022 Jan 6;29(1):8-10. doi: 10.1016/j.stem.2021.12.006. Cell Stem Cell. 2022. PMID: 34995497
-
Time matters: Human blastoids resemble the sequence of blastocyst development.Cell. 2022 Feb 17;185(4):581-584. doi: 10.1016/j.cell.2022.01.005. Cell. 2022. PMID: 35180387
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

