Identification and expression analysis of the glycosyltransferase GT43 family members in bamboo reveal their potential function in xylan biosynthesis during rapid growth
- PMID: 34856932
- PMCID: PMC8638195
- DOI: 10.1186/s12864-021-08192-y
Identification and expression analysis of the glycosyltransferase GT43 family members in bamboo reveal their potential function in xylan biosynthesis during rapid growth
Abstract
Background: Xylan is one of the most abundant hemicelluloses and can crosslink cellulose and lignin to increase the stability of cell walls. A number of genes encoding glycosyltransferases play vital roles in xylan biosynthesis in plants, such as those of the GT43 family. However, little is known about glycosyltransferases in bamboo, especially woody bamboo which is a good substitute for timber.
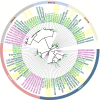
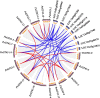
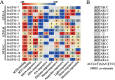
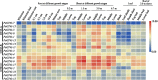
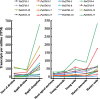
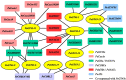
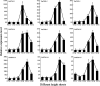
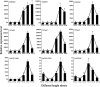
Results: A total of 17 GT43 genes (PeGT43-1 ~ PeGT43-17) were identified in the genome of moso bamboo (Phyllostachys edulis), which belong to three subfamilies with specific motifs. The phylogenetic and collinearity analyses showed that PeGT43s may have undergone gene duplication, as a result of collinearity found in 12 pairs of PeGT43s, and between 17 PeGT43s and 10 OsGT43s. A set of cis-acting elements such as hormones, abiotic stress response and MYB binding elements were found in the promoter of PeGT43s. PeGT43s were expressed differently in 26 tissues, among which the highest expression level was found in the shoots, especially in the rapid elongation zone and nodes. The genes coexpressed with PeGT43s were annotated as associated with polysaccharide metabolism and cell wall biosynthesis. qRT-PCR results showed that the coexpressed genes had similar expression patterns with a significant increase in 4.0 m shoots and a peak in 6.0 m shoots during fast growth. In addition, the xylan content and structural polysaccharide staining intensity in bamboo shoots showed a strong positive correlation with the expression of PeGT43s. Yeast one-hybrid assays demonstrated that PeMYB35 could recognize the 5' UTR/promoter of PeGT43-5 by binding to the SMRE cis-elements.
Conclusions: PeGT43s were found to be adapted to the requirement of xylan biosynthesis during rapid cell elongation and cell wall accumulation, as evidenced by the expression profile of PeGT43s and the rate of xylan accumulation in bamboo shoots. Yeast one-hybrid analysis suggested that PeMYB35 might be involved in xylan biosynthesis by regulating the expression of PeGT43-5 by binding to its 5' UTR/promoter. Our study provides a comprehensive understanding of PeGT43s in moso bamboo and lays a foundation for further functional analysis of PeGT43s for xylan biosynthesis during rapid growth.
Keywords: Glycosyltransferase 43; MYB transcription factors; Phyllostachys edulis; Yeast one-hybrid.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

