The circular RNA expression profile in ovarian serous cystadenocarcinoma reveals a complex circRNA-miRNA regulatory network
- PMID: 34857007
- PMCID: PMC8638095
- DOI: 10.1186/s12920-021-01132-5
The circular RNA expression profile in ovarian serous cystadenocarcinoma reveals a complex circRNA-miRNA regulatory network
Abstract
Background: Ovarian serous cystadenocarcinoma is one of the most serious gynecological malignancies. Circular RNA (circRNA) is a type of noncoding RNA with a covalently closed continuous loop structure. Abnormal circRNA expression might be associated with tumorigenesis because of its complex biological mechanisms by, for example, functioning as a microRNA (miRNA) sponge. However, the circRNA expression profile in ovarian serous cystadenocarcinoma and their associations with other RNAs have not yet been characterized. The main purpose of this study was to reveal the circRNA expression profile in ovarian serous cystadenocarcinoma.
Methods: We collected six specimens from three patients with ovarian serous cystadenocarcinoma and adjacent normal tissues. After RNA sequencing, we analyzed the expression of circRNAs with relevant mRNAs and miRNAs to characterize potential function.
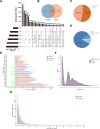
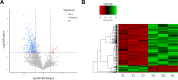
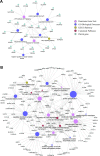
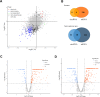
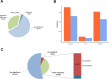
Results: 15,092 unique circRNAs were identified in six specimens. Approximately 46% of these circRNAs were not recorded in public databases. We then reported 353 differentially expressed circRNAs with oncogenes and tumor-suppressor genes. Furthermore, a conjoint analysis with relevant mRNAs revealed consistent changes between circRNAs and their homologous mRNAs. Overall, construction of a circRNA-miRNA network suggested that 4 special circRNAs could be used as potential biomarkers.
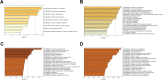
Conclusions: Our study revealed the circRNA expression profile in the tissues of patients with ovarian serous cystadenocarcinoma. The differential expression of circRNAs was thought to be associated with ovarian serous cystadenocarcinoma in the enrichment analysis, and co-expression analysis with relevant mRNAs and miRNAs illustrated the latent regulatory network. We also constructed a complex circRNA-miRNA interaction network and then demonstrated the potential function of certain circRNAs to aid future diagnosis and treatment.
Keywords: Expression profile; Interaction network; Ovarian serous cystadenocarcinoma; circRNA.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018: cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Kawamoto S, Urban BA, Fishman EK. CT of epithelial ovarian tumors. Radio Graph. 1999;19(1):85–102. - PubMed
-
- Bali A, O’Brien PM, Edwards LS, Sutherland RL, Hacker NF, Henshall SM. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin Cancer Res. 2004;10(15):5168–5177. - PubMed
-
- Hefler LA, Zeillinger R, Grimm C, Sood AK, Cheng WF, Gadducci A, et al. Preoperative serum vascular endothelial growth factor as a prognostic parameter in ovarian cancer. Gynecol Oncol. 2006;103(2):512–517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

