Transcriptomic characterization of prurigo nodularis and the therapeutic response to nemolizumab
- PMID: 34857395
- PMCID: PMC8995330
- DOI: 10.1016/j.jaci.2021.10.004
Transcriptomic characterization of prurigo nodularis and the therapeutic response to nemolizumab
Abstract
Background: Prurigo nodularis (PN) is a debilitating, difficult-to-treat, intensely pruritic, chronic inflammatory skin disease characterized by hyperkeratotic skin nodules. The pathogenesis of PN is not well understood but is believed to involve cross talk between sensory nerve fibers, immune cells, and the epidermis. It is centered around the neuroimmune cytokine IL-31, driving an intractable itch-scratch cycle.
Objective: We sought to provide a comprehensive view of the transcriptomic changes in PN skin and characterize the mechanism of action of the anti-IL-31 receptor inhibitor nemolizumab.
Method: RNA sequencing of biopsy samples obtained from a cohort of patients treated with the anti-IL-31 receptor inhibitor nemolizumab and taken at baseline and week 12. Generation and integration of patient data with RNA-Seq data generated from reconstructed human epidermis stimulated with IL-31 and other proinflammatory cytokines.
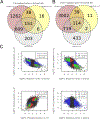
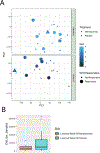
Results: Our results demonstrate that nemolizumab effectively decreases IL-31 responses in PN skin, leading to effective suppression of downstream inflammatory responses including TH2/IL-13 and TH17/IL-17 responses. This is accompanied by decreased keratinocyte proliferation and normalization of epidermal differentiation and function. Furthermore, our results demonstrate how transcriptomic changes associated with nemolizumab treatment correlate with improvement in lesions, pruritus, stabilization of extracellular matrix remodeling, and processes associated with cutaneous nerve function.
Conclusion: These data demonstrate a broad response to IL-31 receptor inhibition with nemolizumab and confirm the critical upstream role of IL-31 in PN pathogenesis.
Keywords: Prurigo nodularis; T(H)17; T(H)2; mechanism; nemolizumab.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Comment in
-
Reversal of the disease signature in prurigo nodularis by blocking the itch cytokine.J Allergy Clin Immunol. 2022 Apr;149(4):1213-1215. doi: 10.1016/j.jaci.2022.02.009. Epub 2022 Feb 18. J Allergy Clin Immunol. 2022. PMID: 35189125 No abstract available.
References
-
- Huang AH, Williams KA, Kwatra SG. Prurigo nodularis: Epidemiology and clinical features. Journal of the American Academy of Dermatology. 2020;83(6):1559–65. - PubMed
-
- Weigelt N, Metze D, Stander S. Prurigo nodularis: systematic analysis of 58 histological criteria in 136 patients. J Cutan Pathol. 2010;37(5):578–86. - PubMed
-
- Johansson O, Liang Y, Marcusson JA, Reimert CM. Eosinophil cationic protein- and eosinophil-derived neurotoxin/eosinophil protein X-immunoreactive eosinophils in prurigo nodularis. Arch Dermatol Res. 2000;292(8):371–8. - PubMed
-
- Liang Y, Marcusson JA, Jacobi HH, Haak-Frendscho M, Johansson O. Histamine-containing mast cells and their relationship to NGFr-immunoreactive nerves in prurigo nodularis: a reappraisal. J Cutan Pathol. 1998;25(4):189–98. - PubMed
-
- Sonkoly E, Muller A, Lauerma AI, Pivarcsi A, Soto H, Kemeny L, et al. IL-31: a new link between T cells and pruritus in atopic skin inflammation. J Allergy Clin Immunol. 2006;117(2):411–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

