Repression of germline genes by PRC1.6 and SETDB1 in the early embryo precedes DNA methylation-mediated silencing
- PMID: 34857746
- PMCID: PMC8639735
- DOI: 10.1038/s41467-021-27345-x
Repression of germline genes by PRC1.6 and SETDB1 in the early embryo precedes DNA methylation-mediated silencing
Abstract
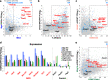
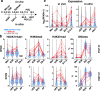
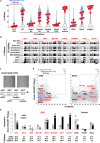
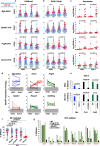
Silencing of a subset of germline genes is dependent upon DNA methylation (DNAme) post-implantation. However, these genes are generally hypomethylated in the blastocyst, implicating alternative repressive pathways before implantation. Indeed, in embryonic stem cells (ESCs), an overlapping set of genes, including germline "genome-defence" (GGD) genes, are upregulated following deletion of the H3K9 methyltransferase SETDB1 or subunits of the non-canonical PRC1 complex PRC1.6. Here, we show that in pre-implantation embryos and naïve ESCs (nESCs), hypomethylated promoters of germline genes bound by the PRC1.6 DNA-binding subunits MGA/MAX/E2F6 are enriched for RING1B-dependent H2AK119ub1 and H3K9me3. Accordingly, repression of these genes in nESCs shows a greater dependence on PRC1.6 than DNAme. In contrast, GGD genes are hypermethylated in epiblast-like cells (EpiLCs) and their silencing is dependent upon SETDB1, PRC1.6/RING1B and DNAme, with H3K9me3 and DNAme establishment dependent upon MGA binding. Thus, GGD genes are initially repressed by PRC1.6, with DNAme subsequently engaged in post-implantation embryos.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
DNMTs and SETDB1 function as co-repressors in MAX-mediated repression of germ cell-related genes in mouse embryonic stem cells.PLoS One. 2018 Nov 7;13(11):e0205969. doi: 10.1371/journal.pone.0205969. eCollection 2018. PLoS One. 2018. PMID: 30403691 Free PMC article.
-
Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities.Mol Cell. 2019 Jun 6;74(5):1037-1052.e7. doi: 10.1016/j.molcel.2019.04.002. Epub 2019 Apr 24. Mol Cell. 2019. PMID: 31029542 Free PMC article.
-
MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6.PLoS Genet. 2018 Jan 30;14(1):e1007193. doi: 10.1371/journal.pgen.1007193. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29381691 Free PMC article.
-
Polycomb-dependent histone H2A ubiquitination links developmental disorders with cancer.Trends Genet. 2022 Apr;38(4):333-352. doi: 10.1016/j.tig.2021.07.011. Epub 2021 Aug 20. Trends Genet. 2022. PMID: 34426021 Review.
-
The functions of SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) in biological process and disease.Epigenetics Chromatin. 2023 Dec 7;16(1):47. doi: 10.1186/s13072-023-00519-1. Epigenetics Chromatin. 2023. PMID: 38057834 Free PMC article. Review.
Cited by
-
Polycomb function in early mouse development.Cell Death Differ. 2025 Jan;32(1):90-99. doi: 10.1038/s41418-024-01340-3. Epub 2024 Jul 12. Cell Death Differ. 2025. PMID: 38997437 Review.
-
Comprehensive profiling of migratory primordial germ cells reveals niche-specific differences in non-canonical Wnt and Nodal-Lefty signaling in anterior vs posterior migrants.bioRxiv [Preprint]. 2025 Jun 2:2024.08.29.610420. doi: 10.1101/2024.08.29.610420. bioRxiv. 2025. PMID: 39257761 Free PMC article. Preprint.
-
Initiation of meiosis from human iPSCs under defined conditions through identification of regulatory factors.Sci Adv. 2025 Aug 15;11(33):eadu0384. doi: 10.1126/sciadv.adu0384. Epub 2025 Aug 15. Sci Adv. 2025. PMID: 40815662 Free PMC article.
-
MGA directly recruits SETDB1/ATF7IP for histone H3K9me3 mark on meiosis-related genes in mouse embryonic stem cells.iScience. 2025 Jul 5;28(8):113059. doi: 10.1016/j.isci.2025.113059. eCollection 2025 Aug 15. iScience. 2025. PMID: 40727931 Free PMC article.
-
Crosstalk within and beyond the Polycomb repressive system.J Cell Biol. 2024 May 6;223(5):e202311021. doi: 10.1083/jcb.202311021. Epub 2024 Mar 20. J Cell Biol. 2024. PMID: 38506728 Free PMC article. Review.
References
-
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. - PubMed
-
- Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019). - PubMed
-
- Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

