Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides
- PMID: 34857750
- PMCID: PMC8639739
- DOI: 10.1038/s41467-021-27144-4
Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides
Abstract
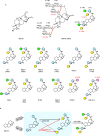
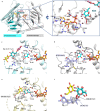
Steviol glycosides are the intensely sweet components of extracts from Stevia rebaudiana. These molecules comprise an invariant steviol aglycone decorated with variable glycans and could widely serve as a low-calorie sweetener. However, the most desirable steviol glycosides Reb D and Reb M, devoid of unpleasant aftertaste, are naturally produced only in trace amounts due to low levels of specific β (1-2) glucosylation in Stevia. Here, we report the biochemical and structural characterization of OsUGT91C1, a glycosyltransferase from Oryza sativa, which is efficient at catalyzing β (1-2) glucosylation. The enzyme's ability to bind steviol glycoside substrate in three modes underlies its flexibility to catalyze β (1-2) glucosylation in two distinct orientations as well as β (1-6) glucosylation. Guided by the structural insights, we engineer this enzyme to enhance the desirable β (1-2) glucosylation, eliminate β (1-6) glucosylation, and obtain a promising catalyst for the industrial production of naturally rare but palatable steviol glycosides.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Enhanced Production of Rebaudioside D and Rebaudioside M through V155T Substitution in the Glycosyltransferase UGT91D2 from Stevia rebaudiana.J Agric Food Chem. 2025 Jan 22;73(3):2019-2032. doi: 10.1021/acs.jafc.4c09392. Epub 2025 Jan 9. J Agric Food Chem. 2025. PMID: 39783863 Free PMC article.
-
Microbial production of next-generation stevia sweeteners.Microb Cell Fact. 2016 Dec 7;15(1):207. doi: 10.1186/s12934-016-0609-1. Microb Cell Fact. 2016. PMID: 27923373 Free PMC article.
-
Mutations in the uridine diphosphate glucosyltransferase 76G1 gene result in different contents of the major steviol glycosides in Stevia rebaudiana.Phytochemistry. 2019 Jun;162:141-147. doi: 10.1016/j.phytochem.2019.03.008. Epub 2019 Mar 18. Phytochemistry. 2019. PMID: 30897351
-
A review on rebaudioside M: The next generation steviol glycoside and noncaloric sweetener.J Food Sci. 2024 Nov;89(11):6946-6965. doi: 10.1111/1750-3841.17401. Epub 2024 Sep 25. J Food Sci. 2024. PMID: 39323262 Review.
-
From Caá-ehé to a commercial sweetener - the diterpenoid glycosides of Stevia rebaudiana.Sci Prog. 2016 Dec 1;99(4):413-419. doi: 10.3184/003685016X14773090197508. Sci Prog. 2016. PMID: 28742480 Free PMC article. Review.
Cited by
-
Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'-O-glycosyltransferase from Scutellaria baicalensis.Acta Pharm Sin B. 2024 Aug;14(8):3746-3759. doi: 10.1016/j.apsb.2024.04.001. Epub 2024 Apr 5. Acta Pharm Sin B. 2024. PMID: 39220864 Free PMC article.
-
Biosynthesis and metabolic engineering of natural sweeteners.AMB Express. 2025 Mar 18;15(1):50. doi: 10.1186/s13568-025-01864-y. AMB Express. 2025. PMID: 40100508 Free PMC article. Review.
-
Enhanced Production of Rebaudioside D and Rebaudioside M through V155T Substitution in the Glycosyltransferase UGT91D2 from Stevia rebaudiana.J Agric Food Chem. 2025 Jan 22;73(3):2019-2032. doi: 10.1021/acs.jafc.4c09392. Epub 2025 Jan 9. J Agric Food Chem. 2025. PMID: 39783863 Free PMC article.
-
Molecular characterization and structural basis of a promiscuous glycosyltransferase for β-(1,6) oligoglucoside chain glycosides biosynthesis.Plant Biotechnol J. 2025 Jun;23(6):2242-2253. doi: 10.1111/pbi.70059. Epub 2025 Mar 19. Plant Biotechnol J. 2025. PMID: 40107321 Free PMC article.
-
Glycosyltransferase engineering and multi-glycosylation routes development facilitating synthesis of high-intensity sweetener mogrosides.iScience. 2022 Sep 27;25(10):105222. doi: 10.1016/j.isci.2022.105222. eCollection 2022 Oct 21. iScience. 2022. PMID: 36248741 Free PMC article.
References
-
- Ahmed SH, Guillem K, Vandaele Y. Sugar addiction: pushing the drug-sugar analogy to the limit. Curr. Opin. Clin. Nutr. Metab. Care. 2013;16:434–439. - PubMed
-
- Editorial. Crystallizing sugar science. Nat. Med. 2016;22:1369. - PubMed
-
- Philippe RN, De Mey M, Anderson J, Ajikumar PK. Biotechnological production of natural zero-calorie sweeteners. Curr. Opin. Biotechnol. 2014;26:155–161. - PubMed
-
- Ceunen S, Geuns JM. Steviol glycosides: chemical diversity, metabolism, and function. J. Nat. Products. 2013;76:1201–1228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

