Dietary excess regulates absorption and surface of gut epithelium through intestinal PPARα
- PMID: 34857752
- PMCID: PMC8639731
- DOI: 10.1038/s41467-021-27133-7
Dietary excess regulates absorption and surface of gut epithelium through intestinal PPARα
Abstract
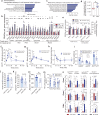
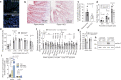
Intestinal surface changes in size and function, but what propels these alterations and what are their metabolic consequences is unknown. Here we report that the food amount is a positive determinant of the gut surface area contributing to an increased absorptive function, reversible by reducing daily food. While several upregulated intestinal energetic pathways are dispensable, the intestinal PPARα is instead necessary for the genetic and environment overeating-induced increase of the gut absorptive capacity. In presence of dietary lipids, intestinal PPARα knock-out or its pharmacological antagonism suppress intestinal crypt expansion and shorten villi in mice and in human intestinal biopsies, diminishing the postprandial triglyceride transport and nutrient uptake. Intestinal PPARα ablation limits systemic lipid absorption and restricts lipid droplet expansion and PLIN2 levels, critical for droplet formation. This improves the lipid metabolism, and reduces body adiposity and liver steatosis, suggesting an alternative target for treating obesity.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

