Fis Connects Two Sensory Pathways, Quorum Sensing and Surface Sensing, to Control Motility in Vibrio parahaemolyticus
- PMID: 34858358
- PMCID: PMC8630636
- DOI: 10.3389/fmicb.2021.669447
Fis Connects Two Sensory Pathways, Quorum Sensing and Surface Sensing, to Control Motility in Vibrio parahaemolyticus
Abstract
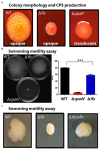
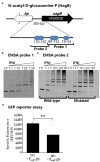
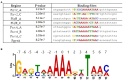
Factor for inversion stimulation (Fis) is a global regulator that is highly expressed during exponential phase growth and undetectable in stationary phase growth. Quorum sensing (QS) is a global regulatory mechanism that controls gene expression in response to changes in cell density and growth phase. In Vibrio parahaemolyticus, a marine species and a significant human pathogen, the QS regulatory sRNAs, Qrr1 to Qrr5, are expressed during exponential growth and negatively regulate the high cell density QS master regulator OpaR. OpaR is a positive regulator of capsule polysaccharide (CPS) formation, which is required for biofilm formation, and is a repressor of lateral flagella required for swarming motility. In V. parahaemolyticus, we show that Fis is a positive regulator of the qrr sRNAs expression. In an in-frame fis deletion mutant, qrr expression was repressed and opaR expression was induced. The Δfis mutant produced CPS and biofilm, but swarming motility was abolished. Also, the fis deletion mutant was more sensitive to polymyxin B. Swarming motility requires expression of both the surface sensing scrABC operon and lateral flagella laf operon. Our data showed that in the Δfis mutant both laf and scrABC genes were repressed. Fis controlled swarming motility indirectly through the QS pathway and directly through the surface sensing pathway. To determine the effects of Fis on cellular metabolism, we performed in vitro growth competition assays, and found that Δfis was outcompeted by wild type in minimal media supplemented with intestinal mucus as a sole nutrient source. The data showed that Fis positively modulated mucus components L-arabinose, D-gluconate and N-acetyl-D-glucosamine catabolism gene expression. In an in vivo colonization competition assay, Δfis was outcompeted by wild type, indicating Fis is required for fitness. Overall, these data demonstrate a global regulatory role for Fis in V. parahaemolyticus that includes QS, motility, and metabolism.
Keywords: Fis; metabolism; motility; quorum sensing; swarming.
Copyright © 2021 Tague, Regmi, Gregory and Boyd.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
LinkOut - more resources
Full Text Sources

