Carbapenem-Resistant Citrobacter spp. as an Emerging Concern in the Hospital-Setting: Results From a Genome-Based Regional Surveillance Study
- PMID: 34858870
- PMCID: PMC8632029
- DOI: 10.3389/fcimb.2021.744431
Carbapenem-Resistant Citrobacter spp. as an Emerging Concern in the Hospital-Setting: Results From a Genome-Based Regional Surveillance Study
Abstract
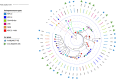
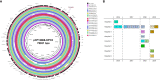
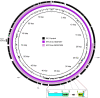
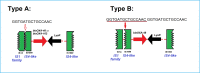
The rise of Carbapenem-resistant Enterobacterales (CRE) represents an increasing threat to patient safety and healthcare systems worldwide. Citrobacter spp., long considered not to be a classical nosocomial pathogen, in contrast to Klebsiella pneumoniae and Escherichia coli, is fast gaining importance as a clinical multidrug-resistant pathogen. We analyzed the genomes of 512 isolates of 21 CRE species obtained from 61 hospitals within a three-year-period and found that Citrobacter spp. (C. freundii, C. portucalensis, C. europaeus, C. koseri and C. braakii) were increasingly detected (n=56) within the study period. The carbapenemase-groups detected in Citrobacter spp. were KPC, OXA-48/-like and MBL (VIM, NDM) accounting for 42%, 31% and 27% respectively, which is comparable to those of K. pneumoniae in the same study. They accounted for 10%, 17% and 14% of all carbapenemase-producing CRE detected in 2017, 2018 and 2019, respectively. The carbapenemase genes were almost exclusively located on plasmids. The high genomic diversity of C. freundii is represented by 22 ST-types. KPC-2 was the predominantly detected carbapenemase (n=19) and was located in 95% of cases on a highly-conserved multiple-drug-resistance-gene-carrying pMLST15 IncN plasmid. KPC-3 was rarely detected and was confined to a clonal outbreak of C. freundii ST18. OXA-48 carbapenemases were located on plasmids of the IncL/M (pOXA-48) type. About 50% of VIM-1 was located on different IncN plasmids (pMLST7, pMLST5). These results underline the increasing importance of the Citrobacter species as emerging carriers of carbapenemases and therefore as potential disseminators of Carbapenem- and multidrug-resistance in the hospital setting.
Keywords: ARGs; Carbapenemase; Citrobacter; Germany; IncN-plasmid; MLST; WGS.
Copyright © 2021 Yao, Falgenhauer, Falgenhauer, Hauri, Heinmüller, Domann, Chakraborty and Imirzalioglu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Arana D. M., Ortega A., González-Barberá E., Lara N., Bautista V., Gómez-Ruíz D., et al. (2017. a). Carbapenem-Resistant Citrobacter Spp. Isolated in Spain From 2013 to 2015 Produced a Variety of Carbapenemases Including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J. Antimicrob. Chemother. 72, 3283–3287. doi: 10.1093/jac/dkx325 - DOI - PubMed
-
- Arana D. M., Ortega A., González-Barberá E., Lara N., Bautista V., Gómez-Ruíz D., et al. (2017. b). Carbapenem-Resistant Citrobacter Spp. Isolated in Spain From 2013 to 2015 Produced a Variety of Carbapenemases Including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J. Antimicrob. Chemother. 72, 3283–3287. doi: 10.1093/jac/dkx325 - DOI - PubMed
-
- Babiker A., Evans D. R., Griffith M. P., McElheny C. L., Hassan M., Clarke L. G., et al. (2020). Clinical and Genomic Epidemiology of Carbapenem- Nonsusceptible Citrobacter Spp. At a Tertiary Health Care Center Over 2 Decades. J. Clin. Microbiol. 58 (9), e00275–20. doi: 10.1128/JCM.00275-20 - DOI - PMC - PubMed
-
- Brolund A., Lagerqvist N., Byfors S., Struelens M. J., Monnet D. L., Albiger B., et al. (2019). Worsening Epidemiological Situation of Carbapenemase-Producing Enterobacteriaceae in Europe, Assessment by National Experts From 37 Countries, July 2018. Eurosurveillance 24 (9), 1900123. doi: 10.2807/1560-7917.ES.2019.24.9.1900123 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

