Variant interpretation using population databases: Lessons from gnomAD
- PMID: 34859531
- PMCID: PMC9160216
- DOI: 10.1002/humu.24309
Variant interpretation using population databases: Lessons from gnomAD
Abstract
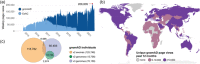
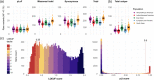
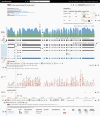
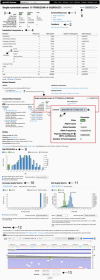
Reference population databases are an essential tool in variant and gene interpretation. Their use guides the identification of pathogenic variants amidst the sea of benign variation present in every human genome, and supports the discovery of new disease-gene relationships. The Genome Aggregation Database (gnomAD) is currently the largest and most widely used publicly available collection of population variation from harmonized sequencing data. The data is available through the online gnomAD browser (https://gnomad.broadinstitute.org/) that enables rapid and intuitive variant analysis. This review provides guidance on the content of the gnomAD browser, and its usage for variant and gene interpretation. We introduce key features including allele frequency, per-base expression levels, constraint scores, and variant co-occurrence, alongside guidance on how to use these in analysis, with a focus on the interpretation of candidate variants and novel genes in rare disease.
Keywords: allele frequency; constraint; database; gnomAD; reference population; variant interpretation.
© 2022 The Authors. Human Mutation Published by Wiley Periodicals LLC.
Conflict of interest statement
D.G.M. is a founder with equity of Goldfinch Bio, and serves as a paid advisor to GSK, Variant Bio, Insitro, and Foresite Labs. A.O.‐D.L. is on the Scientific Advisory Board for Congenica.
Figures
References
-
- Abou Tayoun, A. N. , Pesaran, T. , DiStefano, M. T. , Oza, A. , Rehm, H. L. , & Biesecker, L. G. , ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) . (2018). Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Human Mutation, 39(11), 1517–1524. - PMC - PubMed
-
- Van der Auwera, G. A. , Carneiro, M. O. , Hartl, C. , Poplin, R. , Del Angel, G. , Levy‐Moonshine, A. , DePristo, … , & M. A. (2013). From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics/Editoral Board, Andreas D. Baxevanis … [et Al.], 43, 11.10.1–11.10.33. - PMC - PubMed
-
- Carlston, C. M. , O'Donnell‐Luria, A. H. , Underhill, H. R. , Cummings, B. B. , Weisburd, B. , Minikel, E. V. , & Mao, R. (2017). Pathogenic ASXL1 somatic variants in reference databases complicate germline variant interpretation for Bohring‐Opitz Syndrome. Human Mutation, 38(5), 517–523. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

