ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data
- PMID: 34860150
- PMCID: PMC8767331
- DOI: 10.1099/mgen.0.000728
ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data
Abstract
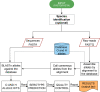
Escherichia coli is a priority foodborne pathogen of public health concern and phenotypic serotyping provides critical information for surveillance and outbreak detection activities. Public health and food safety laboratories are increasingly adopting whole-genome sequencing (WGS) for characterizing pathogens, but it is imperative to maintain serotype designations in order to minimize disruptions to existing public health workflows. Multiple in silico tools have been developed for predicting serotypes from WGS data, including SRST2, SerotypeFinder and EToKi EBEis, but these tools were not designed with the specific requirements of diagnostic laboratories, which include: speciation, input data flexibility (fasta/fastq), quality control information and easily interpretable results. To address these specific requirements, we developed ECTyper (https://github.com/phac-nml/ecoli_serotyping) for performing both speciation within Escherichia and Shigella, and in silico serotype prediction. We compared the serotype prediction performance of each tool on a newly sequenced panel of 185 isolates with confirmed phenotypic serotype information. We found that all tools were highly concordant, with 92-97 % for O-antigens and 98-100 % for H-antigens, and ECTyper having the highest rate of concordance. We extended the benchmarking to a large panel of 6954 publicly available E. coli genomes to assess the performance of the tools on a more diverse dataset. On the public data, there was a considerable drop in concordance, with 75-91 % for O-antigens and 62-90 % for H-antigens, and ECTyper and SerotypeFinder being the most concordant. This study highlights that in silico predictions show high concordance with phenotypic serotyping results, but there are notable differences in tool performance. ECTyper provides highly accurate and sensitive in silico serotype predictions, in addition to speciation, and is designed to be easily incorporated into bioinformatic workflows.
Keywords: E. coli; enteric pathogens; in silico serotyping; public health; serotyping.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
-
- World Health Organization WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference groupestimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. World Health Organization; 2015.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

