Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome
- PMID: 34861039
- PMCID: PMC8972092
- DOI: 10.1182/blood.2021012652
Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome
Abstract
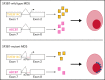
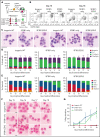
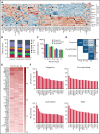
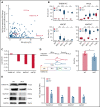
SF3B1 splicing factor mutations are near-universally found in myelodysplastic syndromes (MDS) with ring sideroblasts (RS), a clonal hematopoietic disorder characterized by abnormal erythroid cells with iron-loaded mitochondria. Despite this remarkably strong genotype-to-phenotype correlation, the mechanism by which mutant SF3B1 dysregulates iron metabolism to cause RS remains unclear due to an absence of physiological models of RS formation. Here, we report an induced pluripotent stem cell model of SF3B1-mutant MDS that for the first time recapitulates robust RS formation during in vitro erythroid differentiation. Mutant SF3B1 induces missplicing of ∼100 genes throughout erythroid differentiation, including proposed RS driver genes TMEM14C, PPOX, and ABCB7. All 3 missplicing events reduce protein expression, notably occurring via 5' UTR alteration, and reduced translation efficiency for TMEM14C. Functional rescue of TMEM14C and ABCB7, but not the non-rate-limiting enzyme PPOX, markedly decreased RS, and their combined rescue nearly abolished RS formation. Our study demonstrates that coordinated missplicing of mitochondrial transporters TMEM14C and ABCB7 by mutant SF3B1 sequesters iron in mitochondria, causing RS formation.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures
Comment in
-
RNA missplicing and ring sideroblasts in MDS.Blood. 2022 Mar 31;139(13):1933-1935. doi: 10.1182/blood.2021014923. Blood. 2022. PMID: 35357481 No abstract available.
References
-
- Cazzola M, Malcovati L. Diagnosis and treatment of sideroblastic anemias: from defective heme synthesis to abnormal RNA splicing. Hematology. 2015;2015:19-25. - PubMed
-
- Darman RB, Seiler M, Agrawal AA, et al. . Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033-1045. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA015704/CA/NCI NIH HHS/United States
- S10 OD028685/OD/NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R21 HL139864/HL/NHLBI NIH HHS/United States
- R01 CA251138/CA/NCI NIH HHS/United States
- R01 DK103854/DK/NIDDK NIH HHS/United States
- R01 HL031823/HL/NHLBI NIH HHS/United States
- R00 HL123484/HL/NHLBI NIH HHS/United States
- T32 GM008268/GM/NIGMS NIH HHS/United States
- DP2 HL147126/HL/NHLBI NIH HHS/United States
- T32 HL007093/HL/NHLBI NIH HHS/United States
- R01 HL128239/HL/NHLBI NIH HHS/United States
- M01 RR000037/RR/NCRR NIH HHS/United States
- R35 GM119835/GM/NIGMS NIH HHS/United States
- R01 HL151651/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

