The chromosome-level genome provides insight into the molecular mechanism underlying the tortuous-branch phenotype of Prunus mume
- PMID: 34861048
- PMCID: PMC9299681
- DOI: 10.1111/nph.17894
The chromosome-level genome provides insight into the molecular mechanism underlying the tortuous-branch phenotype of Prunus mume
Abstract
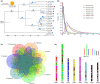
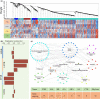
Plant with naturally twisted branches is referred to as a tortuous-branch plant, which have extremely high ornamental value due to their zigzag shape and the natural twisting of their branches. Prunus mume is an important woody ornamental plant. However, the molecular mechanism underlying this unique trait in Prunus genus is unknown. Here, we present a chromosome-level genome assembly of the cultivated P. mume var. tortuosa created using Oxford Nanopore combined with Hi-C scaffolding, which resulted in a 237.8 Mb genome assembly being anchored onto eight pseudochromosomes. Molecular dating indicated that P. mume is the most recently differentiated species in Prunus. Genes associated with cell division, development and plant hormones play essential roles in the formation of tortuous branch trait. A putative regulatory pathway for the tortuous branch trait was constructed based on gene expression levels. Furthermore, after transferring candidate PmCYCD genes into Arabidopsis thaliana, we found that seedlings overexpressing these genes exhibited curled rosette leaves. Our results provide insights into the evolutionary history of recently differentiated species in Prunus genus, the molecular basis of stem morphology, and the molecular mechanism underlying the tortuous branch trait and highlight the utility of multi-omics in deciphering the properties of P. mume plant architecture.
Keywords: Prunus mume; genome assembly; molecular mechanism; plant architecture; tortuous branch.
© 2021 The Authors New Phytologist © 2021 New Phytologist Foundation.
Figures



Comment in
-
Decoding the molecular regulation mechanism of plant architecture in woody plants.New Phytol. 2022 Jul;235(1):8-10. doi: 10.1111/nph.18145. Epub 2022 Apr 23. New Phytol. 2022. PMID: 35460568 No abstract available.
References
-
- Alioto T, Alexiou KG, Bardil A, Barteri F, Castanera R, Cruz F, Dhingra A, Duval H, Fernández i Martí Á, Frias L et al. 2020. Transposons played a major role in the diversification between the closely related almond and peach genomes: results from the almond genome sequence. The Plant Journal 101: 455–472. - PMC - PubMed
-
- Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local aligment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

