Retroviral RNase H: Structure, mechanism, and inhibition
- PMID: 34861939
- PMCID: PMC8994160
- DOI: 10.1016/bs.enz.2021.07.007
Retroviral RNase H: Structure, mechanism, and inhibition
Abstract
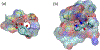
All retroviruses encode the enzyme, reverse transcriptase (RT), which is involved in the conversion of the single-stranded viral RNA genome into double-stranded DNA. RT is a multifunctional enzyme and exhibits DNA polymerase and ribonuclease H (RNH) activities, both of which are essential to the reverse-transcription process. Despite the successful development of polymerase-targeting antiviral drugs over the last three decades, no bona fide inhibitor against the RNH activity of HIV-1 RT has progressed to clinical evaluation. In this review article, we describe the retroviral RNH function and inhibition, with primary consideration of the structural aspects of inhibition.
Keywords: Dynamics; HIV-1; Retrovirus; Reverse transcriptase; Ribonuclease H; Structure.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures

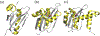
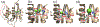

References
-
- Coffin JM, Hughes SH, Varmus HE, Retroviruses, Cold Spring Harbor Laboratory Press, Plainview, NY, 1997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
