Evidence of unidirectional gene flow in a fragmented population of Salmo trutta L
- PMID: 34862454
- PMCID: PMC8642411
- DOI: 10.1038/s41598-021-02975-9
Evidence of unidirectional gene flow in a fragmented population of Salmo trutta L
Abstract
Selection, genetic drift, and gene flow affect genetic variation within populations and genetic differences among populations. Both drift and selection tend to decrease variation within populations and increase differences among populations, whereas gene flow increases variation within populations but leads to populations being related. In brown trout (Salmo trutta L.), the most important factor in population fragmentation is disrupted river-segment connectivity. The main goal of the study was to use genetic analysis to estimate the level of gene flow among resident and migratory brown trout in potential hybridization areas located downstream of impassable barriers in one river basin in the southern Baltic Sea region. First, spawning redds were counted in the upper river basin downstream of impassable barriers. Next, samples were collected from juveniles in spawning areas located downstream of barriers and from adults downstream and upstream of barriers. Subsequently, genetic analysis was performed using a panel of 13 microsatellite loci and the Salmo trutta 5 K SNP microarray. The genetic differentiation estimated between the resident form sampled upstream of the barriers and the anadromous specimens downstream of the barriers was high and significant. Analysis revealed that gene flow occurred between the two forms in the hybridization zone investigated and that isolated resident specimens shared spawning grounds with sea trout downstream of the barriers. The brown trout population from the river system investigated was slightly, internally diversified in the area accessible to migration. Simultaneously, the isolated part of the population was very different from that in the rest of the basin. The spawning areas of the anadromous form located downstream of the barriers were in a hybridization zone and gene flow was confirmed to be unidirectional. Although they constituted a small percentage, the genotypes typical upstream of the barriers were admixed downstream of them. The lack of genotypes noted upstream of the barriers among adult anadromous individuals might indicate that migrants of upstream origin and hybrids preferred residency.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






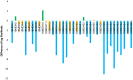
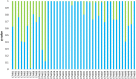

References
-
- Klemetsen A, et al. Atlantic salmon Salmo salar L., brown trout Salmo trutta L. and Arctic charr Salvelinus alpinus (L.): A review of aspects of their life histories. Ecol. Freshw. Fish. 2003;12:1–59. doi: 10.1034/j.1600-0633.2003.00010.x. - DOI
-
- Elliott JM. Quantitative Ecology and the Brown Trout. Oxford University Press; 1994.
-
- ICES Baltic Salmon and Trout Assessment Working Group (WGBAST) ICES Sci. Rep. 2020;2(22):261. doi: 10.17895/ices.pub.5974. - DOI
-
- Berrebi P, Horvath Á, Splendiani A, Palm S, Bernaś R. Genetic diversity of domestic brown trout stocks in Europe. Aquaculture. 2021;544:737043. doi: 10.1016/j.aquaculture.2021.737043. - DOI
-
- Jonsson B, Jonsson N. Partial migration: Niche shift versus sexual maturation in fishes. Rev. Fish Biol. Fish. 1993;3:348–365. doi: 10.1007/BF00043384. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

