HLA-EPI: A new EPIsode in exploring donor/recipient epitopic compatibilities
- PMID: 34862850
- PMCID: PMC9545700
- DOI: 10.1111/tan.14505
HLA-EPI: A new EPIsode in exploring donor/recipient epitopic compatibilities
Abstract
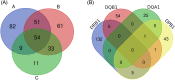
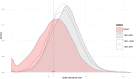
The HLA system plays a pivotal role both in transplantation and immunology. While classical HLA genotypes matching is made at the allelic level, recent progresses were developed to explore antibody-antigen recognition by studying epitopes. Donor to recipient matching at the epitopic level is becoming a trending topic in the transplantation research field because anti-HLA antibodies are epitope-specific rather than allele-specific. Indeed, different HLA alleles often share common epitopes. We present the HLA-Epi tool (hla.univ-nantes.fr) to study an HLA genotype at the epitope level. Using the international HLA epitope registry (Epregistry.com.br) as a reference, we developed HLA-Epi to easily determine epitopic and allelic compatibility levels between several HLA genotypes. The epitope database covers the most common HLA alleles (N = 2976 HLA alleles), representing more than 99% of the total observed frequency of HLA alleles. The freely accessible web tool HLA-Epi calculates an epitopic mismatch load between different sets of potential recipient-donor pairs at different resolution levels. We have characterized the epitopic mismatches distribution in a cohort of more than 10,000 kidney transplanted pairs from European ancestry, which showed low number of epitopic mismatches: 56.9 incompatibilities on average. HLA-Epi allows the exploration of epitope pairing matching to better understand epitopes contribution to immune responses regulation, particularly during transplantation. This free and ready-to-use bioinformatics tool not only addresses limitations of other related tools, but also offers a cost-efficient and reproducible strategy to analyze HLA epitopes as an alternative to HLA allele compatibility. In the future, this could improve sensitization prevention for allograft allocation decisions and reduce the risk of alloreactivity.
Keywords: HLA; HLA epitope; HLA eplet; HSCT; epitopic compatibility; solid organ transplantation.
© 2021 The Authors. HLA: Immune Response Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- European Directorate for the Quality of Medicines and Health Care. International figures on donation and transplantation 2020. Newsletter Transplant; 2021.
-
- Loiseau P, Busson M, Balere ML, et al. HLA association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA‐A, ‐B, ‐C, ‐DRB1, or ‐DQB1 is strongly associated with overall survival. J Am Soc Blood Marrow Transplant. 2007;13(8):965‐974. doi:10.1016/j.bbmt.2007.04.010 - DOI - PubMed
-
- Osorio‐Jaramillo E, Haasnoot GW, Kaider A, et al. HLA epitope mismatching is associated with rejection and worsened graft survival in heart transplant recipients. J Heart Lung Transplant. 2019;38(4, Supplement):S87. doi:10.1016/j.healun.2019.01.199 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

