Epigenome-wide association studies: current knowledge, strategies and recommendations
- PMID: 34863305
- PMCID: PMC8645110
- DOI: 10.1186/s13148-021-01200-8
Epigenome-wide association studies: current knowledge, strategies and recommendations
Abstract
The aetiology and pathophysiology of complex diseases are driven by the interaction between genetic and environmental factors. The variability in risk and outcomes in these diseases are incompletely explained by genetics or environmental risk factors individually. Therefore, researchers are now exploring the epigenome, a biological interface at which genetics and the environment can interact. There is a growing body of evidence supporting the role of epigenetic mechanisms in complex disease pathophysiology. Epigenome-wide association studies (EWASes) investigate the association between a phenotype and epigenetic variants, most commonly DNA methylation. The decreasing cost of measuring epigenome-wide methylation and the increasing accessibility of bioinformatic pipelines have contributed to the rise in EWASes published in recent years. Here, we review the current literature on these EWASes and provide further recommendations and strategies for successfully conducting them. We have constrained our review to studies using methylation data as this is the most studied epigenetic mechanism; microarray-based data as whole-genome bisulphite sequencing remains prohibitively expensive for most laboratories; and blood-based studies due to the non-invasiveness of peripheral blood collection and availability of archived DNA, as well as the accessibility of publicly available blood-cell-based methylation data. Further, we address multiple novel areas of EWAS analysis that have not been covered in previous reviews: (1) longitudinal study designs, (2) the chip analysis methylation pipeline (ChAMP), (3) differentially methylated region (DMR) identification paradigms, (4) methylation quantitative trait loci (methQTL) analysis, (5) methylation age analysis and (6) identifying cell-specific differential methylation from mixed cell data using statistical deconvolution.
Keywords: Bioinformatics; ChAMP; Complex diseases; EWAS; Epigenetics; Methylation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
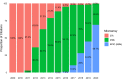








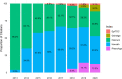


References
-
- Zheleznyakova GY, Piket E, Marabita F, et al. Epigenetic research in multiple sclerosis: progress, challenges, and opportunities. Physiol Genomics. 2017;49(9):447–461. - PubMed
-
- Li X, Xiao B, Chen X-S. DNA methylation: a new player in multiple sclerosis. Mol Neurobiol. 2017;54(6):4049–4059. - PubMed
-
- Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics. 2009;1(1):177–200. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

