Multiparameter immunohistochemistry analysis of HIV DNA, RNA and immune checkpoints in lymph node tissue
- PMID: 34863818
- PMCID: PMC9036546
- DOI: 10.1016/j.jim.2021.113198
Multiparameter immunohistochemistry analysis of HIV DNA, RNA and immune checkpoints in lymph node tissue
Abstract
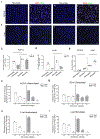
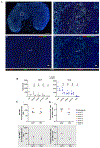
The main barrier to a cure for HIV is the persistence of long-lived and proliferating latently infected CD4+ T-cells despite antiretroviral therapy (ART). Latency is well characterized in multiple CD4+ T-cell subsets, however, the contribution of regulatory T-cells (Tregs) expressing FoxP3 as well as immune checkpoints (ICs) PD-1 and CTLA-4 as targets for productive and latent HIV infection in people living with HIV on suppressive ART is less well defined. We used multiplex detection of HIV DNA and RNA with immunohistochemistry (mIHC) on formalin-fixed paraffin embedded (FFPE) cells to simultaneously detect HIV RNA and DNA and cellular markers. HIV DNA and RNA were detected by in situ hybridization (ISH) (RNA/DNAscope) and IHC was used to detect cellular markers (CD4, PD-1, FoxP3, and CTLA-4) by incorporating the tyramide system amplification (TSA) system. We evaluated latently infected cell lines, a primary cell model of HIV latency and excisional lymph node (LN) biopsies collected from people living with HIV (PLWH) on and off ART. We clearly detected infected cells that coexpressed HIV RNA and DNA (active replication) and DNA only (latently infected cells) in combination with IHC markers in the in vitro infection model as well as LN tissue from PLWH both on and off ART. Combining ISH targeting HIV RNA and DNA with IHC provides a platform to detect and quantify HIV persistence within cells identified by multiple markers in tissue samples from PLWH on ART or to study HIV latency.
Keywords: HIV latency; Immune checkpoints; Immunohistochemistry; In situ hybridization; Lymph nodes; Microscopy; RNAscope.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
No conflict of interest to declare.
Figures



References
-
- Aguzzi A, Krautler NJ, 2010. Characterizing follicular dendritic cells: a progress report. Eur. J. Immunol 40, 2134–2138. - PubMed
-
- Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux J-M, de Leval L, Pantaleo G, Perreau M, 2016. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med 22, 754–761. - PubMed
-
- Brooks DG, Hamer DH, Arlen PA, Gao L, Bristol G, Kitchen CMR, Berger EA, Zack JA, 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19, 413–423. - PubMed
-
- Chun T-W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF, 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med 1, 1284–1290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

